In the Northern Hemisphere, winter ushers in the cold and flu season. People with runny noses and sore throats are often debating if they have the flu, "just a cold", or now COVID-19. Since the emergence of COVID-19, there has been increased strain on the health care system during flu season, with emergency room visits and hospitalizations reaching critical numbers. The increased spotlight on respiratory viruses has made Influenza, RSV, and COVID-19 household names, and brought molecular testing methods such as PCR testing and rapid antigen testing into the common lexicon.
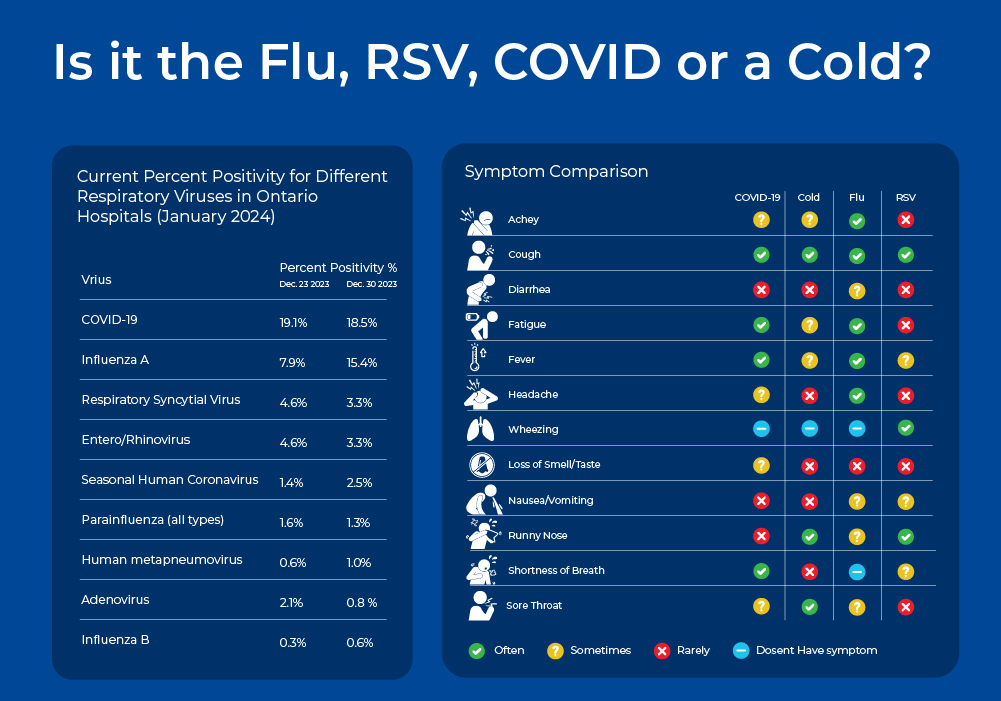
Is it the Flu or Just a Cold?
The common cold, the flu, COVID-19, and RSV are all contagious respiratory illnesses caused by different viruses. These diseases have slightly overlapping symptoms, so it is often difficult to identify the viral culprit without molecular testing. In this blog, we will explore the differences between these viruses in terms of their symptoms, epidemiology, as well as their virion and genome structure which ultimately impact their detection methods. We will also discuss molecular detection methods, as quick and accurate viral identification is important to help with critical bed management decisions in the hospital setting.
Pathogen/Viral Nucleic Acid Isolation Kit (Magnetic Bead System)
For the extraction of nucleic acid (DNA and RNA) from viruses and bacteria found in body fluids and transport mediums. See related application note for supporting clinical data.
Learn More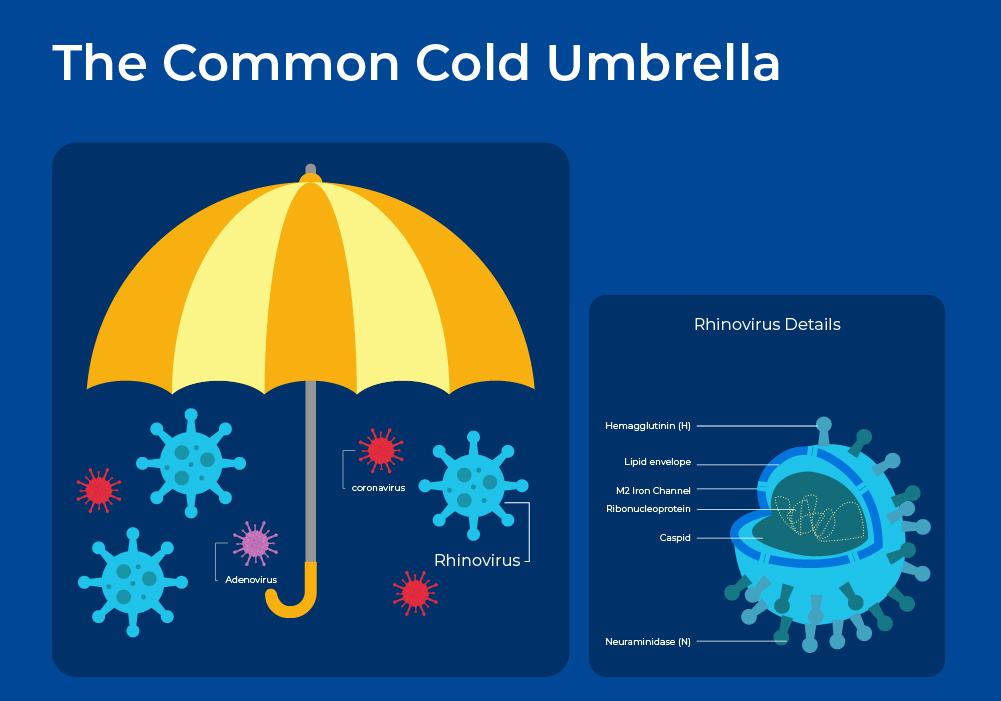
What is the Common Cold?
The common cold is a mild respiratory illness that affects people all over the globe. On average, adults have 2 or 3 colds a year, whereas children can have up to 5 colds a year. The common cold is an umbrella term for over 200 different respiratory virus strains. This viral diversity partially explains the diversity of symptoms experienced with colds, while the host's immune response and comorbidities, such as COPD, further explain the symptom variance in individual patients. Transmission of cold viruses is via airborne droplets, or more often, hand to hand contact with subsequent passage to the nostrils and eyes.
Rhinoviruses are the most prevalent cold-causing virus, followed by coronaviruses. Frequently more than one virus is detectable, but often the infecting organism cannot be identified. Rhinoviruses are named from the Greek "rhinos", meaning "of the nose". This name relates to the fact that rhinoviruses replicate best at the temperatures found in the nose (33-35°C).
Rhinoviruses are positive-sense, single-stranded RNA viruses belonging to the genus Enterovirus. Phylogenetically, human rhinoviruses can be grouped into three species: A, B, and C. Each of these species includes a wide array of serotypes, which differ from each other by variations in their surface proteins. With more than 150 identified serotypes, rhinoviruses collectively contribute to 30-50% of all colds. Rhinovirus A (~ 80 serotypes) and C ( > 50 serotypes) viruses exhibit a stronger association with significant illness and wheezing, whereas rhinovirus B (~ 20 serotypes) species are typically more mild or asymptomatic. The 3 strains, and even minor groups, bind to different human receptors and thus can elicit different immune and inflammatory responses.1
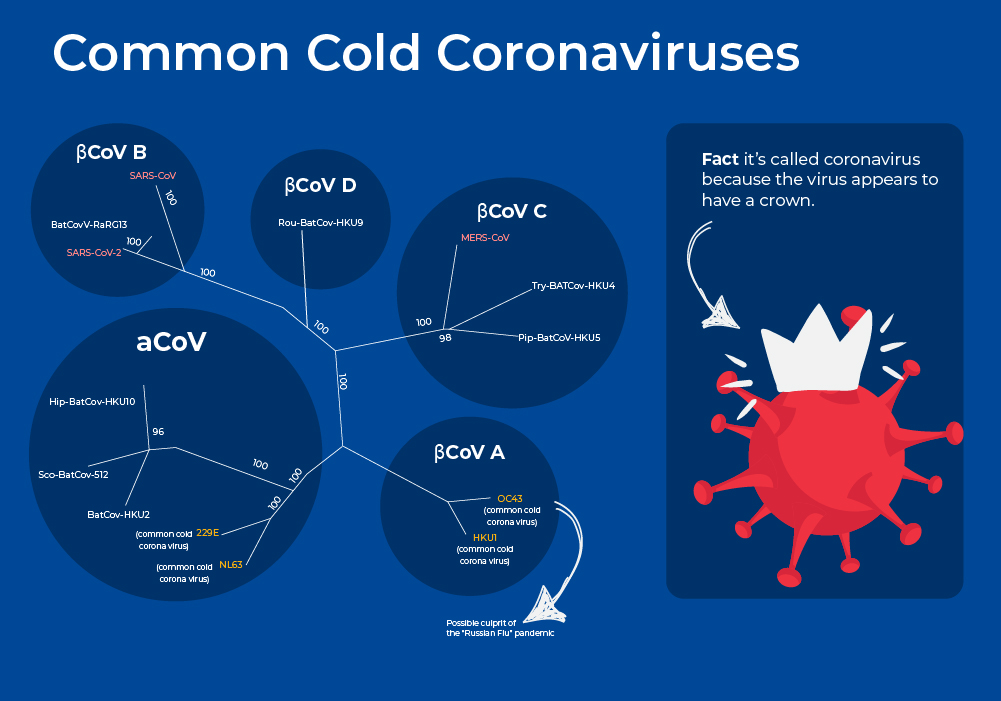
Common Cold Coronaviruses
Coronaviruses are a large family of RNA viruses that, until the 20th century, were mainly known for causing diseases in livestock. In the 1960s, researchers working at the Common Cold Unit of the British Medical Research Council collected a unique common cold virus designated B814 that could not be cultivated using standard techniques which had successfully cultivated rhinoviruses, adenoviruses, and other known common cold viruses. Five years later it was cultivated using a novel technique involving serially passing it through organ culture of human embryonic trachea. Subsequently, two other similar viruses were isolated using organ culture techniques, and all three viruses were shown to be morphologically related by their general shape and distinctive club-like spikes, and thus named "coronavirus", derived from Latin corona, meaning "crown" or "wreath".
Coronaviruses are enveloped viruses with a nucleocapsid of helical symmetry and a positive-sense single-stranded RNA genome. The genome size is one of the largest among RNA viruses, ranging from 26 to 32 kilobases. Pre-pandemic, these four strains had been estimated to be responsible for up to 15%-30% of annual respiratory tract infections, with infections occurring most frequently in young children. Common cold coronaviruses belong to two genera, α-coronaviruses (HCoV-229E and HCoV-NL63) and β-coronaviruses (HCoV-OC43 and HCoV-HKU1).2
New interesting research on these four strains now suggests that strain OC43 may have been responsible for the first Coronavirus pandemic. A pandemic known as "The Russian Flu" in 1890 spread quickly by new mass transit such as trains and steamships. Records indicate similar characteristics such as “perversion of taste and smell,” asymptomatic spreading, increased mortality rates in the elderly, a dry cough, and other symptoms.3 Moreover, sequencing and molecular clock estimates place the emergence of strain OC43 around the same time frame around 1890.4, 5
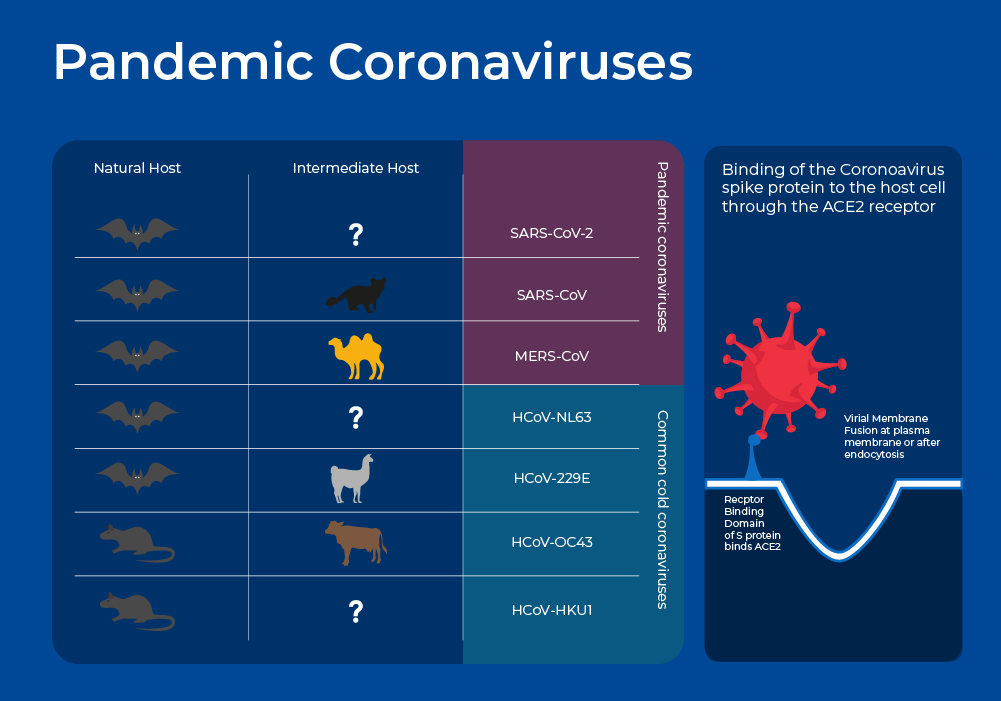
Pandemic Coronaviruses
Global concern surrounding Coronaviruses as virulent human pathogens began in the early 21st century with the epidemic of severe acute respiratory syndrome (SARS) in 2002-2003, and Middle East respiratory syndrome (MERS) in 2012. These new Coronaviruses have much higher case fatality rates than the common cold coronaviruses at 10% and 35%. In late 2019, another new strain emerged - SARS-CoV-2 - and although it has a lower case fatality rate, ~2 %, it is more transmissible and has thus resulted in far more deaths, with almost 7 Million COVID-19 deaths being reported to the World Health Organization6, and many more excess deaths that may be directly or indirectly attributed to COVID-19.
Like other coronaviruses, SARS-CoV-2, SARS, and MERS have four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins. The N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope. The spike protein forms the characteristic crown-like structures on the viral surface and plays an important role in the initiation of the viral cycle as it binds to the ACE2 receptors of the host cell, which then enables the delivery of the viral genome into the cells. This receptor, present in all mammal species, is located at the outer surface of different human cell types, including alveolar cells in the lung, enterocytes in the small intestine, arterial and venous endothelial cells and arterial smooth muscle cells in most organs.4
The spike protein is also important as the vaccines are based on getting our body to produce antibodies against the spike protein. The mRNA vaccines inject mRNA into the muscle's cell. Once inside, they use the cells' machinery to produce a piece of what is called the spike protein.
Although it is generally agreed upon that SARS and MERS are of zoonotic origin and were transferred to humans through an intermediate host, the origin of SARS-CoV-2 is much more murky. At the moment, there is insufficient sequencing data of closely related coronaviruses to identify an intermediate host. Moreover, doubts about the zoonotic origin of SARS-CoV-2 were raised following the observation of four insertions of short sequences within the S protein. The fourth insertion is the most noteworthy. It is a mutation in the furin cleave site that is unique among all the coronaviruses of the SARS group, and is key to its pathogenesis.7 Although similar mutations in the furin site have been noted in other natural viruses, some suggest that this one appears to indicate genetic engineering. The lab origin theory is based on the fact that gain-of-function (GOF) experiments on coronaviruses were conducted on B-coronaviruses in Wuhan China. In gain-of-function experiments, researchers genetically alter organisms to enhance their pathogenicity, thereby gaining a greater understanding of the molecular determinants of virulence.8 There is evidence supporting 9 a natural origin and a laboratory-based origin of the SARS-CoV-2 virus, and further research needs to be conducted to get closer to a definitive answer.

Respiratory syncytial virus (RSV)
Respiratory syncytial virus, or human orthopneumovirus, is a negative-sense, single-stranded RNA virus. Its name is derived from the large cells known as syncytia that form when infected cells fuse. Respiratory syncytial virus (RSV) is a leading cause of lower respiratory tract infection in young children, however reinfection remains common in later life, though often with less severity. The majority of children in North America experience at least one RSV infection before the age of two. RSV infections are usually self-limiting with typical upper respiratory tract symptoms. However, approximately 15-50% of children will go on to develop more serious lower respiratory tract infections, such as bronchiolitis, viral pneumonia, or croup, and up to 80% of hospitalized children with respiratory infections test positive for RSV.
The COVID-19 pandemic disrupted RSV and other respiratory viral infection patterns for the 2021 and 2022 winter with unusually early and high rates of hospitalisations with RSV infections, particularly in 2022. Recent research shows that this was due to Covid in both direct and indirect methods. Non-pharmaceutical interventions such as masking and social distancing prevented RSV from spreading, and built a susceptible population with diminished immunity.10 This "immunity-debt" in children who were not exposed to the virus alone cannot completely explain the substantial increase observed in November 2022. Studies indicate that long-lasting effects of COVID-19 on multiple organ systems, including immune, respiratory, endocrine, cardiovascular, and neurological, have contributed to the surge and severity of pediatric RSV disease.11
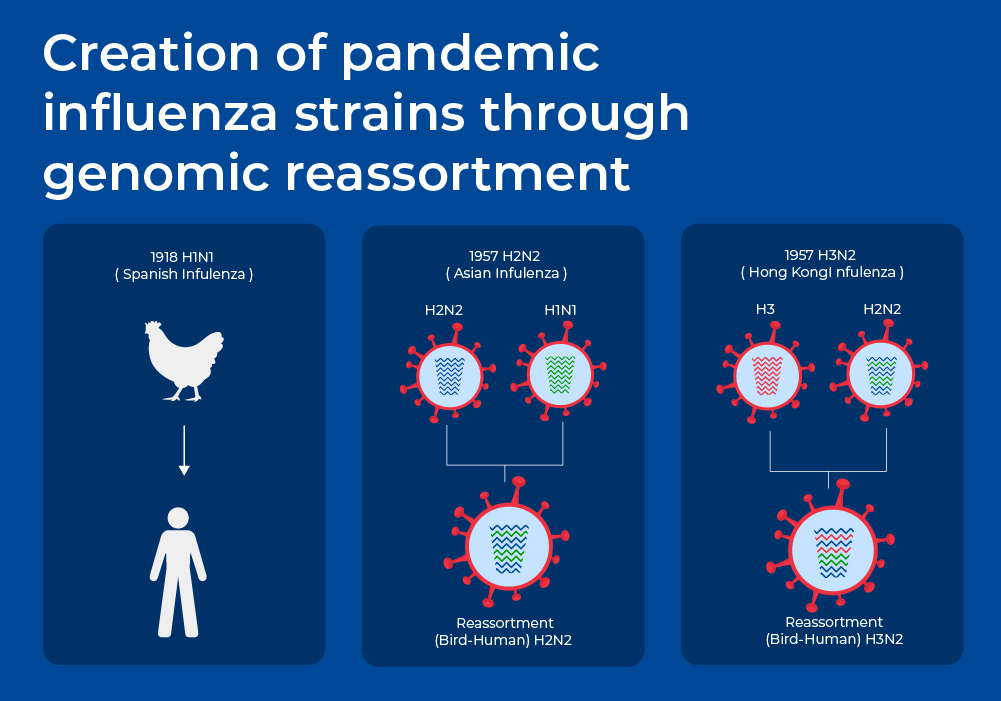
Influenza Viruses
Influenza has a long history with human populations, with frequent seasonal outbreaks, and more occasionally, deadly pandemics. In the northern and southern hemispheres there is a resurgence of the flu every winter, driven by mutations in the genome that allow it to evade antibodies generated in response to the previous flu strain. Periodic outbreaks occur when an influenza strain emerges with significantly different surface proteins, leading to a higher mortality rate. However, survivors develop some immunity and eventually the virus transitions back into the regular seasonal flu, persisting until the advent of the next pandemic flu.
Phylogenetically there are four types of influenza viruses that differ based on their host range, genome structure, and surface proteins. Influenza A and B are the main species circulating in humans. Influenza C virus is detected less frequently and usually causes mild infections, and thus does not present public health importance. Most pandemics have been caused by Influenza A viruses.
Influenza A viruses are further classified into subtypes according to the combination of the proteins on the surface of the virus - hemagglutinin (HA), and neuraminidase (NA). These HA and NA surface proteins are "antigens", that work together to enter host cells. They are recognized by the immune system and are capable of eliciting an immune response, including the production of antibodies. There are 18 types of HA proteins, with the first three hemagglutinins, H1, H2, and H3, found in human influenza viruses. There are 11 subtypes of Neuraminidase. Influenza viruses are named as H1N1, H5N2 etc., depending on the combinations of HA and NA.
Vaccines against the flu are trivalent or quadrivalent, providing protection against an H1N1 strain, an H3N2 strain, and one or two InfluenzaB strains. The WHO's Global Influenza Surveillance and Response System (GISRS) gathers global information of which influenza strains are circulating. Every year the composition of the flu vaccines are adjusted to match the prevailing strains of the current season. One of the reasons that the flu vaccines have to be adjusted so often is that the influenza virus has high mutation rates.12 The influenza RNA-polymerase has very low fidelity, so while the viruses are replicating inside the host's cells, there is little proofreading, and there are slightly different influenza viruses being generated inside each person. In addition to this antigenic drift, the segmented genome structure of the influenza virus means that there can be larger "antigenic shifts" when there are events such as genomic reassortment. For example, some of the influenza pandemics of the past occurred when genome segments from two different species combined. This is presumed to happen occasionally when two different strains infect the same host cells. These usually seem to be combinations with strains of avian origin.13
Molecular Detection of Respiratory Viruses
Acute respiratory tract infections (RTIs) impose a significant disease burden and rank as the third leading cause of global mortality. Among individuals admitted to hospitals with acute RTIs, respiratory viruses emerge as the primary causative factor, constituting 50%-66% of cases.14 Correct and rapid virus identification can improve patient management by influencing decisions surrounding antiviral therapy, antibiotic treatment, hospital admission, quarantining measures, and infection-control measures. It may also lead to avoidance of unnecessary costs and antimicrobial resistance by reducing unnecessary prescriptions of antibiotics.
Many hospital and clinical settings have now transitioned from conventional techniques such as viral cultures and immunoassays, to molecular techniques such as reverse-transcription polymerase chain reaction (RT-PCR) techniques. A recent meta-analysis showed that on average these molecular tests have a sensitivity of 90.9% and a specificity of 96.1%, with some multiplex real-time RT-PCR assays having a sensitivity of 98-99%. Research investigating the clinical implications of employing molecular tests revealed notable reductions in turnaround times, shortened hospital stays, increased appropriate use of antiviral drugs, and a potential decrease in costs and extra radiographs. The majority of high-quality studies demonstrated these benefits when compared to traditional molecular methods. However, no discernible impact was observed on antibiotic prescriptions, or the number of hospital admissions.14
For accurate molecular detection a complete workflow is needed with proper sample preservation, a nucleic acid extraction system that produces an inhibitor-free eluate with high yield, and accurate RT-qPCR kits.