What is COP28
The annual United Nations Climate Change Conference - COP28 UAE, is currently taking place in Dubai. Over 70,000 world leaders, politicians, activists, students and Oil-Company CEOs from across the world are gathering to discuss strategies for addressing climate change. Delegates have met annually for almost three decades to try and find ways to reduce greenhouse gas emissions in an equitable way, as well as to support countries that have been disproportionately affected by climate change.
These meetings have swung between fractious and uneventful, 2015 was viewed as a triumph when the "Paris Agreement" was signed, while Copenhagen 2009 was a disaster, as the conference broke down and delegates went home after the rich countries refused to back a commitment to limit global warming. This year kicked off with some drama as there were questions surrounding the choice of the COP28 President and possible conflict of interest. The COP28 President, Sultan al-Jaber, is also the chief executive of UAE's national oil company, Adnoc. Under his leadership they are planning to increase production from 3 million barrels of oil a day in 2016 to 5 million by 2030. However, many that have worked with him say that he is honestly committed to transitioning, and we need input from the oil and gas industry. Under Al Jaber, Adnoc has also invested in carbon capture and green hydrogen projects, while also committing to power its operations with renewable energy sources. It is obvious that real change requires the input of the Oil and gas industry, so let's remain hopeful that this conference can produce meaningful results.
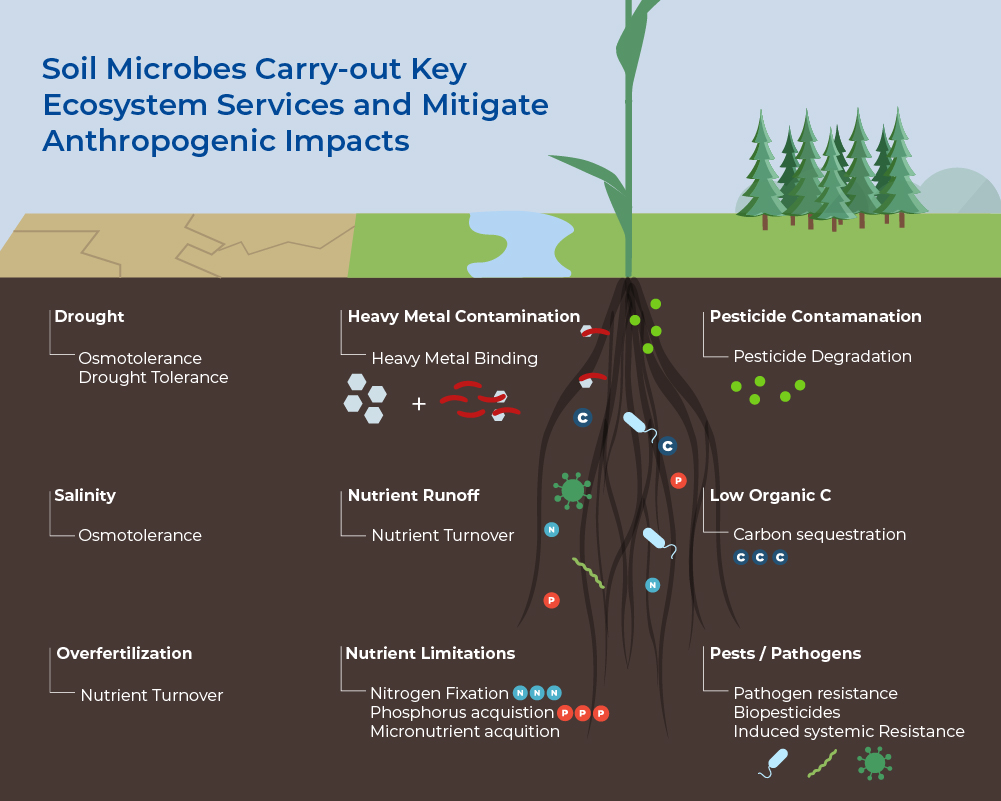
Soil Microbes Carry-out Key Ecosystem Services
One teaspoon of soil contains between 100 million to 1 billion individual bacteria. Despite their size, after plants, microbes in the soil represent the largest fraction of biomass on Earth. Bacteria are most abundant at 15% of the total living biomass, but the biomass of fungi (2%) and Archaea (1%) are also larger than that of animals (0.3%). Soils also harbour the most diverse and complex microbiome on Earth, with often more than 50,000 species per gram. Not only do soil microbes comprise the majority of biological diversity on Earth, but they are the foundation for soil food webs, and therefore underpin the diversity of higher trophic levels. These microbiomes have great metabolic diversity with many enzymatic functions still being unknown. Their combined metabolic potential means that their activities drive, or contribute to, the cycling of all major elements (e.g. C, N, P). Moreover, the nutrient cycling affects the structure and function of the soil ecosystem and provides important Ecosystem Services. Microbes are important for the biological control of pests, weeds, and pathogens, therefore they are important players in global biodiversity. Lastly, they are important players in carbon storage and regulation of greenhouse gas emissions, as both store carbon as stable organic matter and release CO2, CH4 and N2O.1
Want to hear more from Norgen?
Join over 10,000 scientists, bioinformaticians, and researchers who receive our exclusive deals, industry updates, and more, directly to their inbox.
For a limited time, subscribe and SAVE 10% on your next purchase!
SIGN UP
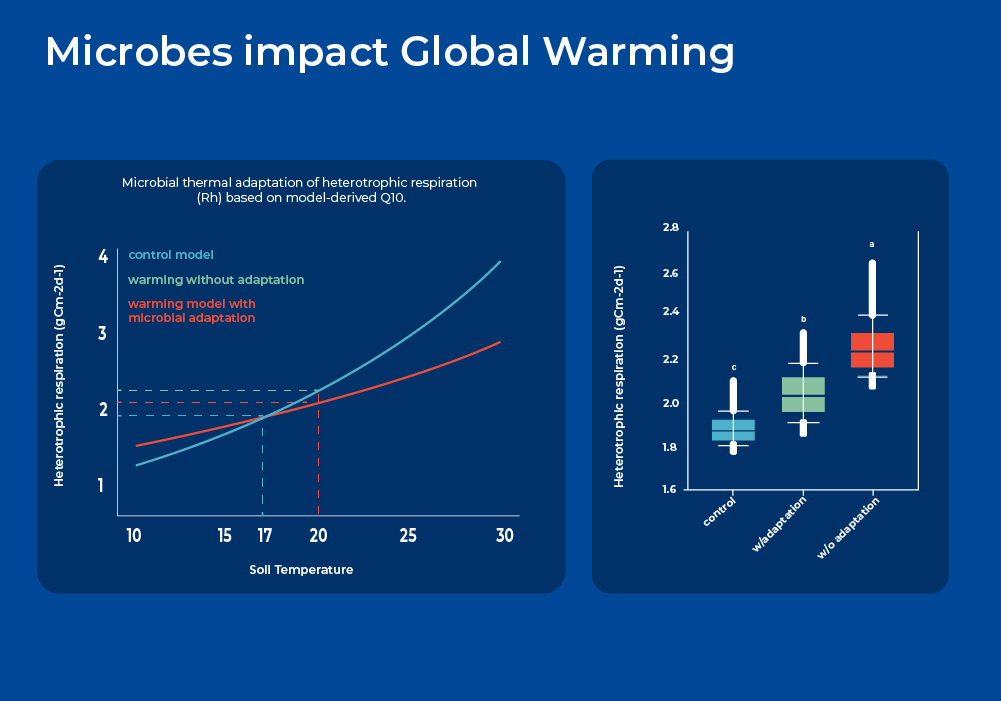
Microbes Impact Global Warming
As mentioned above, microbes in the soil act as both a source and a sink of greenhouse gases as they carry out the dichotomous roles of stabilization of carbon inputs into organic forms, and mineralization of soil organic carbon. On one side, soil stores about three times more organic carbon (C) than the Earth's atmosphere. On the other hand, soil respiration is the largest single source of carbon dioxide (CO2) from terrestrial ecosystems to the atmosphere - about ten times larger than anthropogenic emissions. However, the feedback to climate warming and underlying microbial mechanisms are still poorly understood. Therefore, soil microbial respiration is an important source of uncertainty in projecting future climate and carbon (C) cycle feedbacks. Soil total respiration includes both autotrophic respiration from plant root growth and root biomass maintenance, and heterotrophic respiration from microbial decomposition of litter and soil organic matter (SOM). Various short-term experiments show that soil respiration increases exponentially with temperature, which would suggest a very large positive feedback impact. However, more long term experiments are needed as microbial communities are known to shift as part of thermal adaptation.
In a longer term experiment, researchers exposed plots of land to continuous warming for over 7 years. They extracted soil DNA and performed amplicon sequencing (16S and ITS), quantified gene abundances using a geochip, and correlated this data with carbon flux and respiration measurements. They showed that by incorporating microbial functional gene abundance data into a Microbially-enabled Ecosystem Model, they significantly improved the modeling performance of soil microbial respiration and reduced model parametric uncertainty. Moreover, modeling analyses indicated that as the microbial population shifted through thermal adaptation, there is considerably less heterotrophic respiration and hence less soil C loss. If such microbially-mediated dampening effects can be generalized across different temporal and spatial scales, then the potential positive feedback of soil microbial respiration in response to climate warming may be less than previously predicted.2

Climate Change Impacts on Microbes
As shown above, microorganisms can adjust their respiratory responses to temperature over a long period by changing their metabolism and community structure. At the same time, the eco-physiology of microbes is generally expected to be phylogenetically conserved because of the evolutionary history of a given species. Thus, the ecological traits of microorganisms mirror both phylogenetic constraints and environmental acclimation. However, the degree to which these traits are affected by phylogeny and environment remain uncertain.
Researchers looked into this interaction between phylogeny and the environment in another long term climate experiment in which soil plots were exposed to four different climate scenarios for 10 years: 1) control (Cntrl) had ambient temperature and CO2 concentration, 2) elevated temperature by +2°C (eT), 3) elevated CO2 concentration up to 500 ppm (eCO2), and 4) combined CO2 enrichment and warming (eTeCO2). After extracting soil DNA, they performed soil metagenomic analysis combined with stable isotope probing and examined the response of the microbes to these four climate scenarios. Interestingly, they found three growth strategies of bacterial taxa - rapid, intermediate and slow responders - that are phylogenetically conserved. For example, members of class Bacilli and Sphingobacteria are mainly rapid responders. However, these phylogenetic patterns are partially confounded by environmental factors, as the different climate scenarios alter the growth strategies of over 90% of species. The variance for slow responders is primarily explained by climate, whereas the growth of rapid bacterial responders is more influenced by phylogeny. Overall, these results highlight that it is important to understand the phylogenetic composition of the soil microbiome in addition to climatic constraints to be able to predict the growth strategies of soil microorganisms under global change scenarios.3
Soil Microbes Influence Food Security and Agricultural Health
Earth's population is estimated to reach 8 and 9.8 billion by 2025 and 2050, respectively. We therefore need new solutions to be able to feed this growing population. In addition to this exponential population growth, climate change also brings issues of drought, floods, pests, loss of biodiversity, changes in growing seasons, and reduced yields. These impacts will also be compounded by additional anthropogenic inputs, such as the use of synthetic pesticides and fertilizers. One promising approach to sustainably improve crop production and soil health is to harness the beneficial properties of soil microorganisms. Plants and microbial communities have a complex and interdependent relationship, involving various mechanisms that influence ecological interactions. Plants release photosynthates to soil microbes and in return, certain microbial taxa can positively impact plant growth by acquiring and transporting nutrients to the plant through synthesis of phytohormones and production of metabolites that can protect plants from pathogens. Certain soil microorganisms or communities can be used as soil inoculants to promote plant growth by acting as fertilizers or pesticides, or protecting against stress, thereby increasing crop yield while at the same time reducing reliance on harmful chemicals.
One avenue for developing soil microbial inoculants is the development of Synthetic microbial communities (SynComs). However, achieving this goal requires a comprehensive understanding of natural microbial communities, including the genome sequences and their metabolic potentials. In a recent study, researchers used metagenome-assembled genomes (MAGs) from an arid soil community to reconstruct Genome-scale metabolic networks (GSMNs) and identify a minimal community that would promote plant growth. The minimal community they identified preserved important genes associated with Plant Growth-Promoting Traits (PGPTs), including those involved in iron acquisition, EPS production, potassium solubilization, nitrogen fixation, GABA production, and plant growth hormones (IAA-related tryptophan metabolism). They also then integrated metabolic modelling from five important crop plants, including soybean, maize, sorghum, and sugarcane. This allowed for selection of a core set of species that remain beneficial across different hosts, suggesting their importance independent of the specific plant type. By integrating the host into the model, they showed that the hosts primarily provided amino acids, lipids, and coenzymes, while the SymCom, in addition to PGPTs, supplied carbohydrates, esters, amino acids, and aromatic compounds to the hosts. Thus, through this multi-species community-wide GSMNs analysis they gained insight into the metabolic complementarity between bacterial species and host crop plants, moving towards the development of a broadly applicable microbial soil inoculant.4
The Soil Microbiome, Climate Change and Livestock
In addition to crops, soil microbes influence livestock through direct and indirect mechanisms. One interesting study looked into the interaction between soil microbes, climate change, and livestock practices. In addition to increasing temperatures, the use of antibiotics in livestock is also increasing. In 2017 alone, approximately 11 million kg of antibiotics were sold in the United States for livestock use. Up to 90% of these administered antibiotics are excreted onto soils as un-metabolized and biologically active compounds.
A team of researchers collected samples of prairie soil treated with either a high dose, low dose, or no dose of a common livestock antibiotic (Monensin); the soil was subsequently heated at three different temperatures and left to incubate. For each treatment, the team monitored soil respiration, acidity, microbial community composition and function, carbon and nitrogen cycling, and interactions among microbes. Community composition was assessed using a 16S/ITS metabarcoding approach. They found that with rising heat and antibiotic additions, bacterial populations collapsed, allowing fungi to dominate, resulting in fewer total microbes and less microbial diversity overall. Antibiotics alone increased bioavailable carbon and reduced microbial efficiency.5
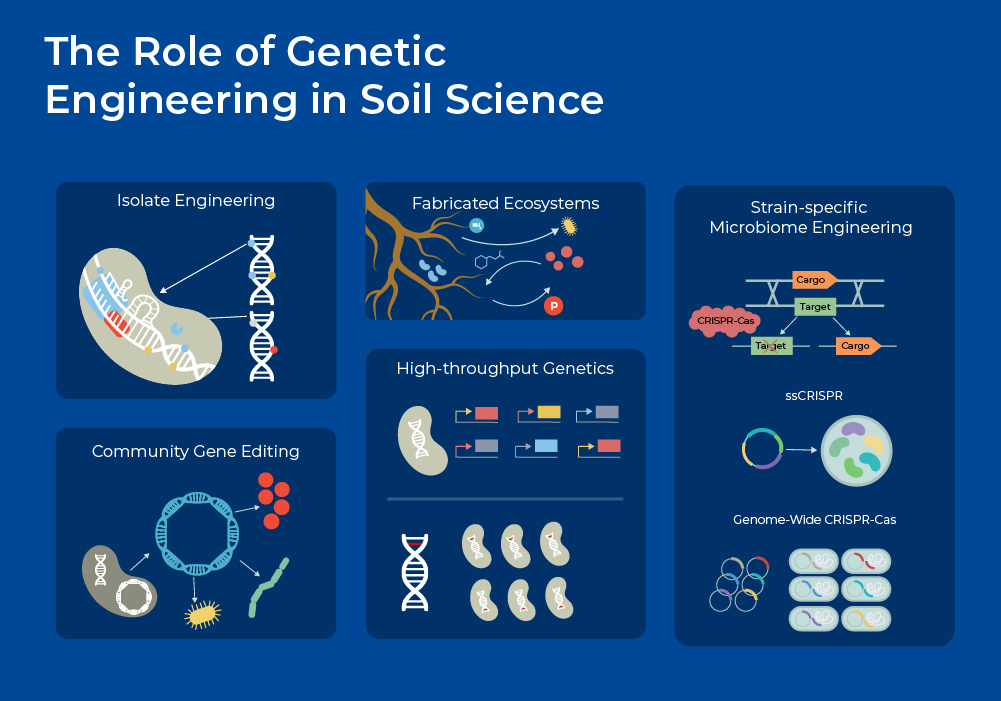
The Role of Genetic Engineering and CRISPR in Soil Science
With advancements in high throughput sequencing, genetic engineering, and culture-independent 'omics, new avenues of soil science are opening up. Researchers are developing new approaches for manipulating the soil microbiome that may mitigate the negative consequences of climate change. Advances in synthetic biology and genetic engineering tools are being used to discover and enhance native microbial functions, introduce new traits, develop biological sensors, and combine multiple beneficial traits into a single organism.1
In one example of engineering soil bacteria, researchers used a combinatorial synthetic biology-based approach to generate a collection of plant-associated bacteria capable of efficient phytate hydrolysis. Microbes that release soluble phosphate from naturally occurring sources in the soil may reduce the need for such fertilizers. They analyzed all available microbial genomes and environmental metagenomes and identified over 2,000 phytate genes across three enzyme classes. They selected 82 of these genes and performed sequence optimization to achieve optimal expression in Proteobacteria. They synthesized these genes, cloned them into high-expression inducible cassettes, and sequence verified them. Finally, they used conjugation to transfer all 82 sequences into the genomes of three root-associated proteobacteria (Pseudomonas simiae, Pseudomonas putida and Ralstonia sp.). Through experiments they demonstrated that these engineered strains improved plant growth under phosphorus-limited conditions. This represents a first step in the development of phosphate-mining bacteria for future use in crop systems and to potentially reduce the reliance on synthetic fertilizers.6
A recent microbiome-wide application of genetic engineering includes in situ microbial community editing using conjugation and CRISPR-Cas technologies. Researchers developed a computational program that can be used to modify microbial consortia through strain-specific CRISPR guide RNAs. They demonstrated the application for isolating a specific strain from the community, as well as for selectively eliminating a specific strain. For strain purification, they utilized guide RNAs (gRNAs) designed to target and kill all microbes in the community except for the specific microbe to be isolated. For strain elimination, they utilized the gRNAs to remove only the unwanted microbe while protecting all other strains. This technique (ssCRISPR) will be of use in diverse microbiota engineering applications, including addressing the problem of antibiotic-resistant microbes and isolating useful microbes from the environment.7
However, questions remain surrounding the ecological impacts of engineered microbes and consortium. These engineered microbes and communities should be tested in Fabricated Ecosystems first to provide community contexts.
Consider Norgen's Soil Related Kits In your next Soil Science Research
Extraction of Nucleic Acids from soil can be an integral part in obtaining a systems level understanding of the soil microbiome. Ultimately, the combination of PCR, NGS, genetic engineering, metabolomics and other techniques will be necessary to meet the challenges that we are facing with climate change and a growing human population.