Multilocus Sequence Typing: An Effective Tool in Studying the Genetic Diversity of Cryptosporidium
What is MLST?
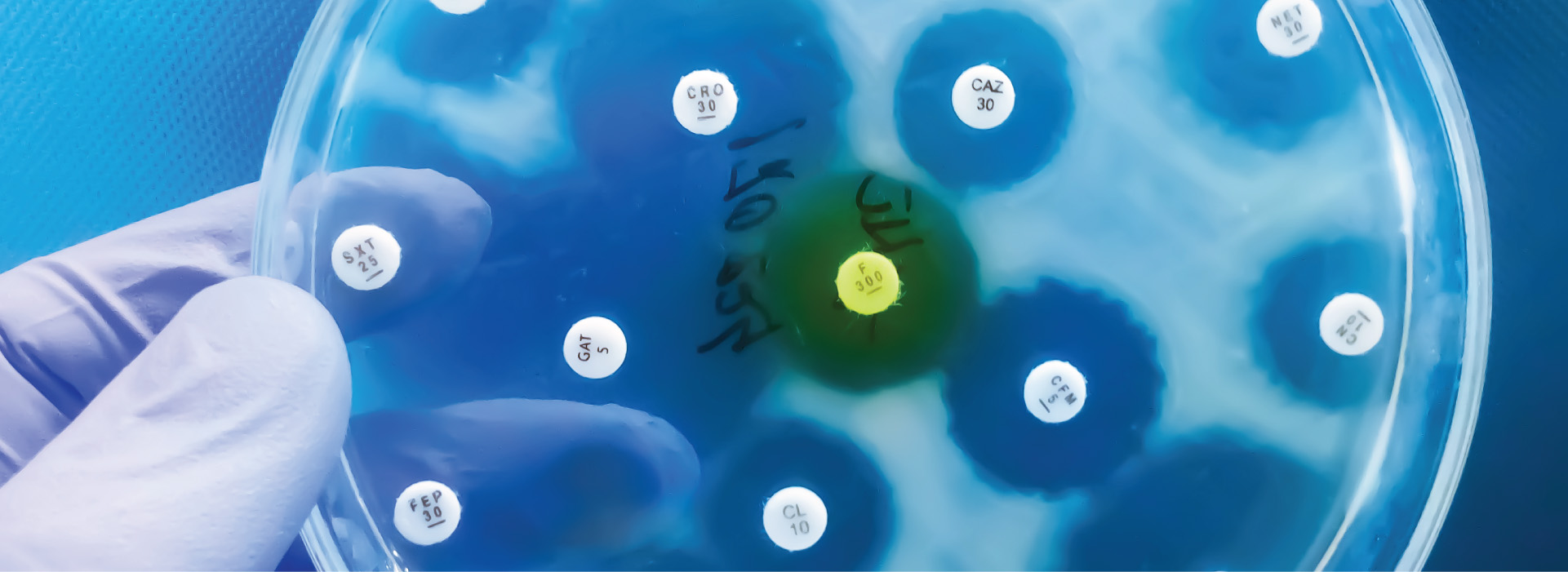
Multilocus sequence typing (MLST) is considered the gold standard method for population analyses of bacteria, fungi, and protists.1 MLST involves the systematic sequencing of established housekeeping genes (usually six to seven) followed by comparison to central databases, enabling unequivocal and standardized characterization of bacterial isolates.1 MLST has been used as a tool to study genetic diversity and transmission dynamics of many clinically relevant bacterial pathogens, including Chlamydia trachomatis and Clostridium difficile.2,3
What is Cryptosporidiosis?
Cryptosporidiosis (commonly known as Crypto) is a highly contagious intestinal infection caused by the parasite Cryptosporidium.4 Symptoms include nausea, abdominal pain, and acute diarrhea. Cryptosporidiosis is transmitted to humans through contact with infected stool, often via contaminated water or food. Cryptosporidiosis is recognized as one of the most common causes of infectious diarrhea worldwide and is associated with high morbidity and mortality.4 As such, it is a disease of great environmental and public health significance.
Studying The Genetic Diversity of Cryptosporidium
In a recent study by Uran-Velasquez et al. (2022), an MLST scheme with seven markers (CP47, MS5, MS9, MSC6-7, TP14, and gp60) was used to analyze the genetic diversity of Cryptosporidium hominis and Cryptosporidium parvum isolated from 28 Colombian patients. Uran-Velasquez et al. (2022) extracted total DNA from patients' stool samples using Norgen Biotek's Stool DNA Isolation Kit (Cat. 27600, 65600). Norgen's kit offers a rapid, convenient methodology for the isolation of high-quality DNA, suitable for sensitive downstream applications such as MLST.
NORBLOG
Want to hear more from Norgen?
Join over 10,000 scientists, bioinformaticians, and researchers who receive our exclusive deals, industry updates, and more, directly to their inbox.
For a limited time, subscribe and SAVE 10% on your next purchase!
SIGN UP
Polymorphic behavior was observed for all seven markers for both C. hominis and C. parvum, especially with the CP47, MS5, and gp60 markers. The gene fixation index (Fst) suggested evolutionary similarity between the C. hominis samples; however, less similarity and greater sequence divergence was observed in the C. parvum samples, offering insight into evolutionary events that have occurred in South American populations of Cryptosporidium.4 Further, their data suggest a predominance of an anthroponotic transmission route. Overall, their study supports the use of MLST as a tool in analyzing the genetic diversity of Cryptosporidium, offering greater depth and detail than could be obtained with unilocular approaches.
-
Page AJ, Alikhan NF, Carleton HA, Seemann T, Keane JA, Katz LS. Comparison of classical multi-locus sequence typing software for next-generation sequencing data. Microb Genom.2017 Jul 4;3(8):e000124.10.1099/mgen.0.000124. PMID: 29026660; PMCID: PMC5610716.
-
Pilo S, Zizelski Valenci G, Rubinstein M, Pichadze L, Scharf Y, Dveyrin Z, Rorman E, Nissan I. High-resolution multilocus sequence typing for Chlamydia trachomatis: improved results for clinical samples with low amounts of C. trachomatis DNA.BMC Microbiol.2021 Jan 18;21(1):28. doi: 10.1186/s12866-020-02077-y. PMID: 33461496; PMCID: PMC7814548.
-
Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE. Multilocus sequence typing of Clostridium difficile.J Clin Microbiol.2010 Mar;48(3):770-8.doi: 10.1128/JCM.01796-09. Epub 2009 Dec 30. PMID: 20042623; PMCID: PMC2832416.
-
Uran-Velasquez J, Alzate JF, Farfan-Garcia AE, Gomez-Duarte OG, Martinez-Rosado LL, Dominguez-Hernandez DD, Rojas W, Galvan-Diaz AL, Garcia-Montoya GM. Multilocus Sequence Typing helps understand the genetic diversity of Cryptosporidium hominis and Cryptosporidium parvum isolated from Colombian patients.PLoS One.2022 Jul 8;17(7):e0270995.doi: 10.1371/journal.pone.0270995. PMID: 35802653; PMCID: PMC9269747.