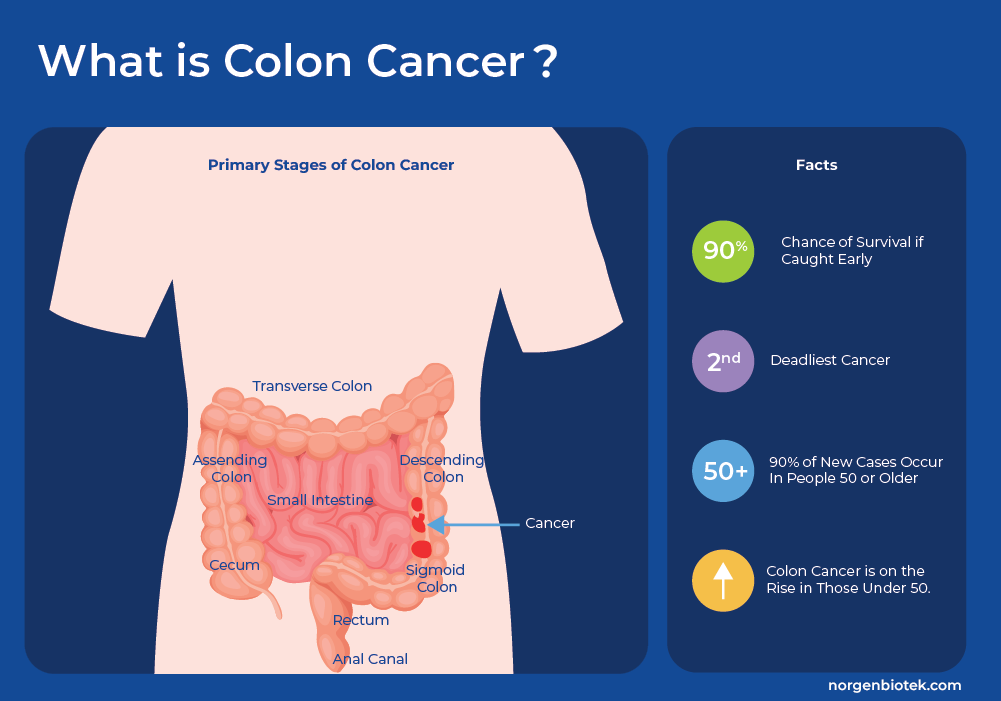
What is Colon Cancer
Colorectal Cancer (CRC), or bowel cancer, which includes colon cancer and rectal cancer, is a disease in which the cells of the colon, or rectum, grow and divide uncontrollably. The colon is the longest part of the large intestine and it connects to the anus through the rectum. Colon cancer starts with the formation of polyps, which are smaller clumps of cells lining the colon. These polyps usually do not cause symptoms, and most often polyps do not progress to cancer. However, occasionally these polyps progress to cancer. There are 5 defined stages of colorectal cancer from Stage 0 to Stage IV as the tumor grows from the inner layer, throughout the colon and possibly throughout the body in final stage IV. If detected early, the chance of survival increases greatly1.
Colorectal cancer is the third most common cancer worldwide, accounting for approximately 10% of all cancer cases and is the second leading cause of cancer-related deaths worldwide.1 It predominantly affects older individuals, with the majority of cases occurring in people aged 50 and above.2 In countries with population-wide screening the overall incidence rates are decreasing. However, recent studies in the US and Europe that analyzed incidence rates by age group, found that the colon cancer risks are increasing among younger adults. Millennials (people born between 1981-1996) have twice the risk of colon cancer compared to those born around 1950, and quadruple the risk of rectal cancer. In the same study, it was shown that individuals under the age of 55 are almost 60% more prone to receiving a late-stage disease diagnosis compared to older adults. That makes the odds of surviving colon cancer more challenging and highlights a need to increase awareness around risk factors and screening in young adults.3, 4
An interesting study on heritability of certain cancers was performed using a cohort of 45,000 twins. It estimated that heredity is not the major risk factor for sporadic cancers. Heritable effects were estimated at 42% for prostate cancer and colon cancer was next at 35%, but this leaves the majority attributed to environmental factors, and chance.5 In addition to family history, significant risk factors for colorectal cancer include inflammatory bowel disease. Lifestyle factors with a statistically significant effect are increased body mass index, smoking cigarettes, and consumption of red meat. Whereas consumption of fruits and vegetables (high-fiber diet) and physical exercise all reduced the risk of colorectal cancer.6
Colon Cancer Screening
When colon cancer is caught early, 9 out of 10 people with the disease can be cured. Therefore, population-wide screening and prevention programs are recommended in many countries. Polyps and colorectal cancer cells can bleed, therefore many screening tests check for small amounts of blood in the stool. At the moment, there are three types of stool tests that are approved by the US Food and Drug Administration (FDA). These tests are normally administered as at-home kits where the patient collects stool at home and then the sample is sent to the doctor.7
- FOBT: The fecal occult blood test (FOBT) test is based on a chemical reaction to detect heme. Heme is a molecular complex that contains iron in the middle and is a central component of red blood cells.
- FIT: The fecal immunochemical test (FIT) is based on antibody detection of hemoglobin. Hemoglobin is a heme-containing protein that is a major component of red blood cells.
- FIT-DNA: Is a combination test that uses antibody detection of hemoglobin combined with detection of specific human DNA sequences that are shed from the intestines into the stool.
Studies have shown that if the FOBT or FIT tests are performed every 1-2 years for people between 50-80 years, it can help reduce the number of deaths.7 Some studies suggest that the FOBT and FIT have lower sensitivity for detection of proximal versus distal CRC due to degradation of the hemoglobin protein 8. Another large clinical study further showed that the FIT-DNA had increased specificity over the FIT test alone, but had more false positive results.9
For individuals with a family history of CRC or who test positive in an at-home stool test, further testing is often prescribed. The most accurate tests in a colonoscopy. A colonoscopy is a procedure in which a long flexible tube with a camera attached, is inserted into the rectum. The video camera allows doctors to visualize any abnormalities in the colon such as polyps or tumors. Although this test is very sensitive, it is also quite invasive, requires special dietary preparations and is often associated with poor adherence as many patients do not want the procedure performed 10. Therefore the development of highly accurate less invasive screening tools is desired.
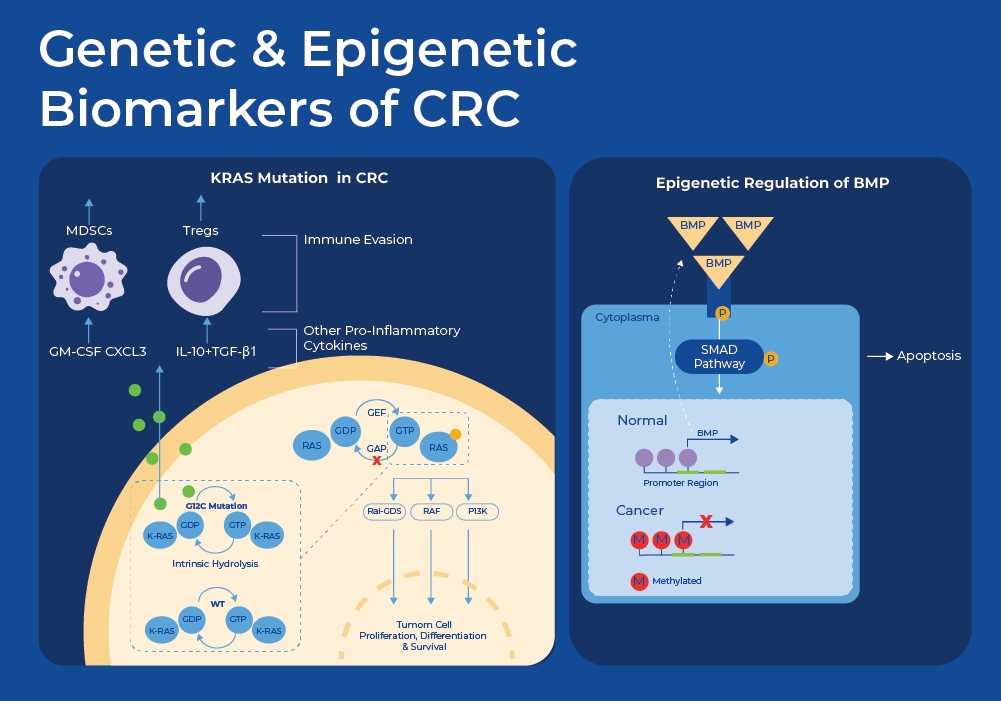
Genetic Mutations as Biomarkers of CRC
At the cellular and molecular level, Colorectal cancer (CRC) is a heterogeneous disease. A wide variety of genetic and epigenetic mutations, transcriptional, and metabolic changes have been identified as biomarkers of CRC. The multi-target FIT-DNA test that is sold by Exact Sciences as Cologuard© includes the detection of a gene mutation known to indicate CRC (KRAS), and two methylated regions (NDRG4, BMP3) plus a hemoglobin immunoassay.
KRAS is a proto-oncogene that is mutated in approximately 40% of all CRC cases11. A proto-oncogene is a normal gene that can become an oncogene due to mutations or increased expression. KRAS encodes a compact protein that binds to guanosine triphosphate (GTP) and guanosine diphosphate (GDP) relaying signals from outside the cell to the cell's nucleus. Its response to extracellular signaling is important for regulation of proliferation and differentiation. Specific mutations result in the constitutive activation of the KRAS protein thus continuously activating these downstream signaling pathways such as cell proliferation, ultimately driving tumorigenesis. Moreover, multiple studies have shown that patients with mutations in KRAS have much poorer prognosis, and these mutations can be used to predict response to treatments. For example mutations in exon 12 (G12V and G12C) were proven to have worse survival rates, as compared to other mutations. Individuals with CRC with mutations in exon 2, exon 3, or exon 4, did not benefit from anti-EGFR therapy, while patients with a mutation in exon 13 (G13D) benefited from chemotherapy plus cetuximab.11
Epigenetic Biomarkers of CRC
Studies are now showing that large-scale epigenetic alterations may yield a better signal for identifying and classifying cancers at an earlier stage than somatic mutations. DNA methylation is one of the best-studied epigenetic mechanisms for the regulation of gene expression. Approximately half of the transcription start sites in the human genome contain CpG islands (CGIs), which are typically unmethylated in somatic cells. Changes in methylation patterns during tumorigenesis have been documented for decades.
SEPT9
The first methylation based screening for Colorectal Cancer that was approved by the US FDA was blood-based quantitative PCR detect methylation of Septin 9 (mSEPT9). Septin 9 (SEPT9) is a gene coding for a GTP-protein that is involved in cytokinesis and cell cycle control. Methylation of this gene is associated with tumorigenesis and is a biomarker for CRC. It was initially approved and marketed as Epi proColon©. Clinical trials and studies have shown specificities ranging from 50% - 90%. A second generation test, Epi proColon 2.0 has now been approved with studies showing specificities and sensitivities around 90%.12
NDG4 and BMP3
The FIT-DNA test commercialized as Cologuard© tests for mutation of KRAS as well as methylation of two genes. They use Quantitative allele-specific real-time target and signal amplification (QuARTS) which combines a polymerase-based target amplification with an invasive cleavage-based signal amplification to quantify methylated BMP3, NDRG4. NDRG4 is a gene belonging to the N-myc downstream regulated gene (NDRG) family. The protein produced by this gene is a cytoplasmic protein essential for cell cycle advancement and survival in primary astrocytes. It potentially participates in regulating mitogenic signaling within vascular smooth muscle cells. NDRG4 plays a role in cell proliferation, apoptosis, and cell differentiation and its methylation status has been associated with multiple cancer types.13 Apoptosis is a specific mechanism of cell death that is used to get rid of abnormal or excess cells. This process is often blocked in cancerous tissue. BMP3, a bone morphogenetic protein, has been studied for its role in tumor progression for nearly three decades. It was Initially identified as a potential tumor suppressor candidate for colorectal cancer (CRC) in 2005 subsequently found to be a target of epigenetic regulation in CRC.
There are many other genes whose epigenetic regulation has been implicated in CRC. Exact Sciences has developed a new panel of highly discriminant candidate methylated DNA markers (MDM). After analyzing methylome-wide sequencing data they identified 12 candidate methylated DNA markers (MDMs) (BMP3, CHST2, CHST10, DMRTA2, LASS4, LRRC4, NDRG4, PDGFD, PPP2R25C, SDC2, SFMBT2, VAV3) 12. In a marker selection experiment using analysis of archived stool samples weighted to early-stage colorectal cancer and advanced precancerous lesions (APL), they showed improved sensitivity and specificity. The new version of the test includes two improvements. It now uses a new reference gene that is constitutively methylated (ZDHHC1). Furthermore, the test no longer detects the KRAS mutation, this allows for using the entire DNA yield in the bisulfite treatment, increasing sensitivity. A multi-center, clinical validation trial is underway.
Other Stool-based Biomarkers

ncRNA as a Biomarker of CRC
In addition to genetic mutations and epigenetic regulation, expression of non-coding RNA (ncRNA) and epigenetic regulation of ncRNA shows signs as a potential biomarker. MicroRNAs (miRNAs) act as post-transcriptional regulators of gene expression by targeting specific genes. miRNAs called oncomirs, that target oncogenes are up-regulated in cancer, while others that silence oncogenes are downregulated in cancer. This impacts the regulation of intracellular signaling networks, inducing cell proliferation, conferring resistance to apoptosis and chemotherapy, and promoting metastasis. A recent meta-analysis identified miR-21 and miR-92a as the most commonly reported fecal-based miRNAs as CRC biomarkers. Many studies have shown that upregulation of miR-21 and miR-92a promotes CRC cell migration, invasion and proliferation and inhibition of apoptosis. There are many reported gene targets of miR-21 that are implicated in colorectal cancer malignancy. The phosphatase and tensin homolog (PTEN) was repeatedly reported to be silenced resulting in the activation of PI3K/AKT pathway and induction of tumor formation. Similarly miR-92a has been shown to interfere with the expression of tumor suppressor genes involved in PI3K/AKT, WNT/Β-catenin and BMP/Smad pathways and enhances tumorigenesis.14
Studies are also showing that dysregulation of miRNAs plays a role in resistance to chemotherapy. Many miRNAs, for example miR-26b and miR-148a, have been shown to have significantly down regulated expression in response to FOLFOX, which is a mixture of folic acid (FOL), 5-fluorouracil (F), and oxaliplatin (OX)15.
Epigenetic Regulation of miRNAs
Epigenetic alterations may occur more frequently than genetic mutations and play a key role in silencing tumor suppressor genes or activating oncogenes, thereby affecting multiple cellular processes. More recently, studies have shown that microRNAs are often dysregulated in colorectal cancer (CRC), via aberrant DNA methylation and that quantifying methylation may be a more sensitive diagnostic biomarker than measuring expression of miRNA.
MiR-124a was one of the first CRC-related microRNAs that was shown to be silenced via epigenetic methylation. Silencing of miR124a by hyper methylation leads to the activation of an oncogene (CDK6) and the phosphorylation of a tumor suppressor gene (retinoblastoma).16 miR-137 is downregulated, through promoter hypermethylation, in many tumors including colorectal cancer. Moreover methylation of miR-137 can be used to differentiate Ulcerative Colitis (UC) patients with high risk of developing CRC. MiR-34, a tumor suppressive microRNA family, has been shown to be directly regulated by the tumor suppressor p53. MiR-34 is frequently methylated in CRC tissues and to a lesser extent in adjacent normal tissues. This suggests that miR-34a-5p is a potential prognostic marker to predict the aggressiveness of cancer in stage II and III CRCs.15
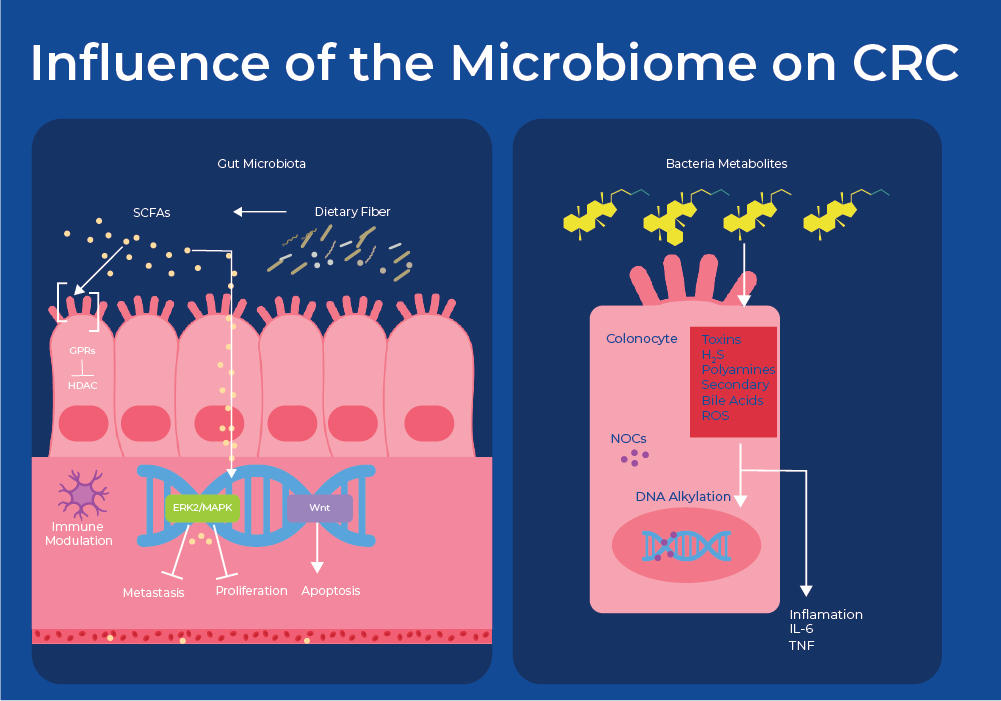
Fecal Metagenomics for CRC
Bacteria can be viewed as causative agents for cancer through mechanisms including chronic inflammation and bacterial production of cancer-promoting or cancer-inhibiting metabolites. For example, Helicobacter pylori is known to cause adenocarcinoma of the distal stomach, or gastric cancer through chronic inflammation. With respect to colon cancer, bacteria metabolize food to produce a variety of molecules. Fermentation of fiber produces short chain fatty acids (SCFA) and metabolism of cholesterol produces secondary bile acids (SBA). Studies have shown that microbial-derived SCFAs control regulatory T cell populations through inhibition of host histone deacetylases (HDAC) and interactions with cell surface receptors. This results in affected cell attachment, immune cell immigration, cytokine production, chemotaxis, and the programmed cell death.17 Moreover, multiple species are involved in the conversion of secondary bile acids into cytotoxic compounds that increase colonic cell proliferation.18 Therefore, an increased abundance of SCFA-producing bacteria is often associated with lower CRC-risk. While an increase in SBA-producing bacteria is often found in patients with CRC. This is supported by the fact that low-fiber and high-fat diets have been shown to be risk factors for CRC.
Zeller et. al (2014) was the first group to examine if microbial biomarkers of CRC can be identified in the gut microbiome. They used metagenomic sequencing of fecal samples to identify taxonomic markers that distinguished CRC patients from tumor-free controls. They created a classifying algorithm that was based on the abundance of 22 microbial species. At least half of the predictive power of the model stemmed from the abundance of the four most discriminative species - two Fusobacterium species, Porphyromonas asaccharolytica and Peptostreptococcus stomatis, all of which are enriched in CRC. Fusobacterium species have been previously shown to accelerate tumorigenesis. Furthermore, some of the shifts in the community can be attributed to metabolic influences. For example, species that efficiently produce butyrate from carbohydrates were consistently depleted in CRC, including R. intestinalis, Roseburia hominis, Anaerostipes hadrus, and Faecalibacterium prausnitzii. Accuracy of metagenomic CRC detection was similar to the standard fecal occult blood test (FOBT) and when both approaches were combined, sensitivity improved > 45% relative to the FOBT, while maintaining its specificity.19
More recently there have been multiple studies that combine fecal metabolomics and metagenomics as a diagnostic tool for colorectal cancer. One group identified bacterial and metabolic differences between early-onset and late-onset colorectal cancer. LO-CRC was characterized by Fusobacterium nucleatum enrichment and short-chain fatty acid depletion, including reduced microbial GABA biosynthesis and a shift in acetate/acetaldehyde metabolism towards acetyl-CoA production. On the other hand, Early onset was associated with Flavonifractor plauti and increased tryptophan, bile acid and choline metabolism. The predictive model based on metagenomic, metabolomic and functional gene markers accurately distinguished EO-CRC from controls.20 Another similar study developed a classification algorithm based on combined metagenomics and metabolism that could distinguish between Colorectal Cancer, Colorectal Adenomas and Healthy Subjects indicating a potential for early diagnosis of colorectal neoplasia.21
VOC-based Detection
Another novel method to screen for Colorectal Cancer involves the detection of volatile organic compounds (VOCs). VOCs are carbon-containing molecules that have a high vapor pressure, a larger proportion of molecules as a gas. VOCs are present in stool or can diffuse into the bloodstream and be passed by the lungs into breath, or into urine through the kidneys. VOCs can be derived from your diet,environment (eg. smoking) or be a product of microbial or human cell metabolism. Neoplasms in the colon are associated with changes in microbial cellular metabolism, and many studies have shown that detection of specific VOCs, or a VOC fingerprint can be used for CRC screening. Examples of some discriminatory compounds include: p-cresol, 3(4H)-DBZ and 1H-indole, p-Cresol 3(4H)-dibenzofuranone, with area under the curve (AUC) values ranging from 70-90%.22 Other groups have used urine-based VOC analysis and identified similar molecules. One found the most discriminatory VOC to be butanol with an AUC of 0.98.23 Urinary VOC analysis does have some advantages over fecal analysis with its ease of collection.
Detection of VOCs can be performed using expensive equipment and personnel or it can be performed using dogs as disease sensors. It has been well established that canines have superior olfactory senses than humans, some reports suggest that their sense of scent is 100,000 times more sensitive than humans. Detection dogs, or sniffer dogs are trained to detect diseases by 'sniffing' or sampling breath, feces, urine, blood, and tissue. Multiple studies have shown that scent-trained dogs can detect COVID-19 in individuals with an 94-96% accuracy and a detection time of 5-10 seconds.24 Interestingly, multiple studies showed that dogs must be re-trained as new variants emerge.25
In an earlier study specific to colon cancer, a trained labrador retriever analyzed breath and stool of CRC patients and healthy controls enrolled in a clinical trial. After smelling the samples, the dog sits down to indicate a positive sample. For breath detection, the sensitivity was 0.91 and the specificity was 0.99. For stool samples, the sensitivity was 0.97 and the specificity was 0.99. The accuracy of canine scent detection was high even for early cancer. Moreover, canine scent detection was not confounded by current smoking, benign colorectal disease or inflammatory disease.26
Norgen Products
Norgen provides an entire workflow starting from preservation of stool through extraction of nucleic acids as well as NGS sequencing. Our preservation kits make sampling easy and keep DNA and RNA stable for 2 years and 7 days respectively. We have many extraction kits for DNA, RNA and Nucleic Acids in a variety of formats (spin columns, 96-well plates and magnetic beads). The preserved stool is also compatible with metabolite analysis. Norgen also offers metagenomic library preparation kits, and associated products.