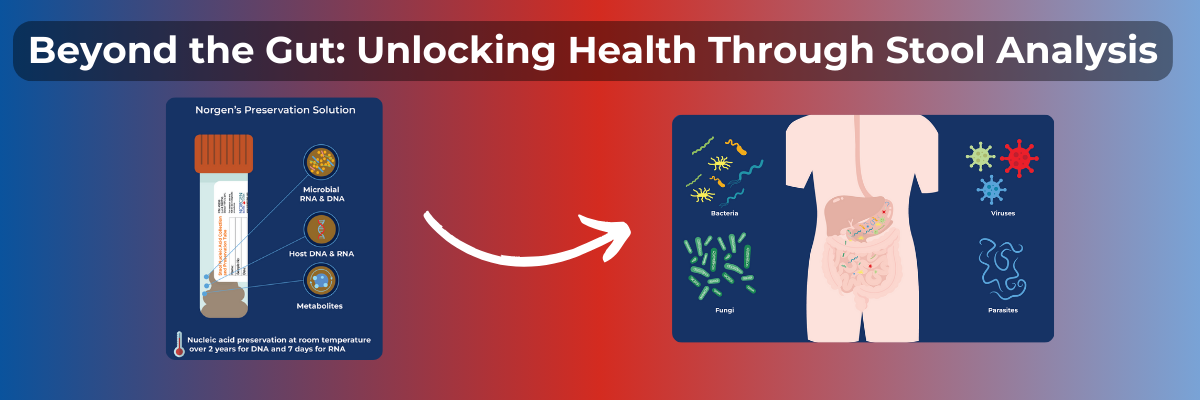
Over the past few years, there has been a lot of talk and numerous studies published surrounding the relationship between the gut microbiota and overall human health. Recently, studies have shown an association between the gut microbiome with obesity, type 2 diabetes, colorectal cancer, and more.1
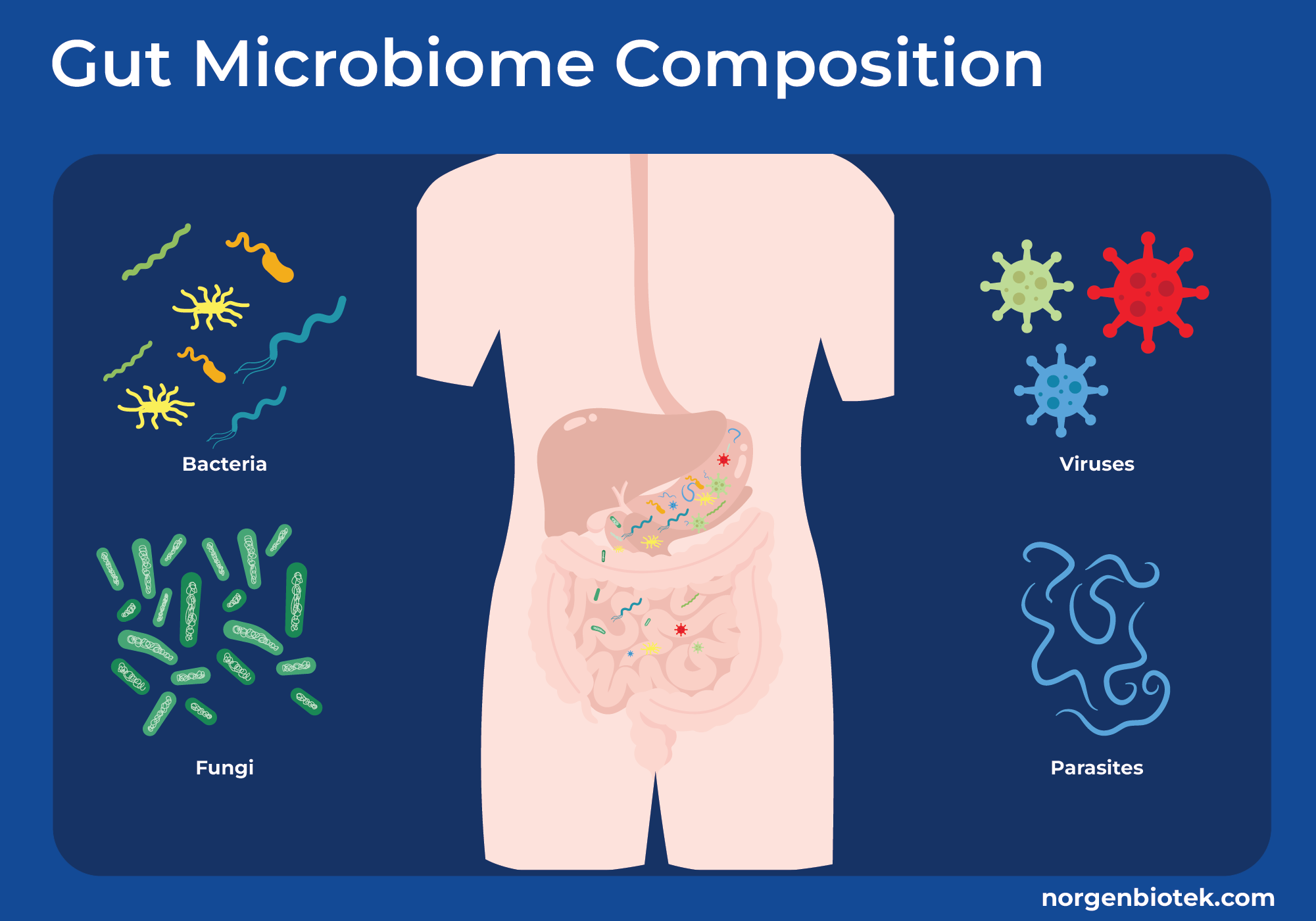
Let's first discuss what the gut microbiome is made of. The gut microbiome is comprised of 10-100 trillion microbial cells, mainly containing over 300 different species of bacteria along with viruses, fungi, and parasites.2, 3, 4 Understanding these microbial cells and their genetic makeup can help us find out about disease correlation, immune system regulation, and metabolomic functions. In this blog, we will discuss how analytes like RNA, DNA, and metabolites from host and microbes can be used to assess the status of human health.
Want to hear more from Norgen?
Join over 10,000 scientists, bioinformaticians, and researchers who receive our exclusive deals, industry updates, and more, directly to their inbox.
For a limited time, subscribe and SAVE 10% on your next purchase!
SIGN UP
Now, where does the investigation of your gut microbes begin? The non-invasive way to investigate the gut is through the stool. The stool RNA and DNA can be used for quantitative PCR analysis, metagenomic sequencing, and even metabolomics. This can not only reveal microbial diversity and antibiotic-resistant genes within the microbiome, it can also aid in the detection of pathogens and diseases. Beginning with stool sample collection, each step of the stool analysis is carefully completed to ensure accurate results. The steps may include stool collection, stool sample preservation, DNA and RNA extraction, and lastly, specific analysis like PCR testing and/or sequencing performed to detect specific microbes and biomarkers.
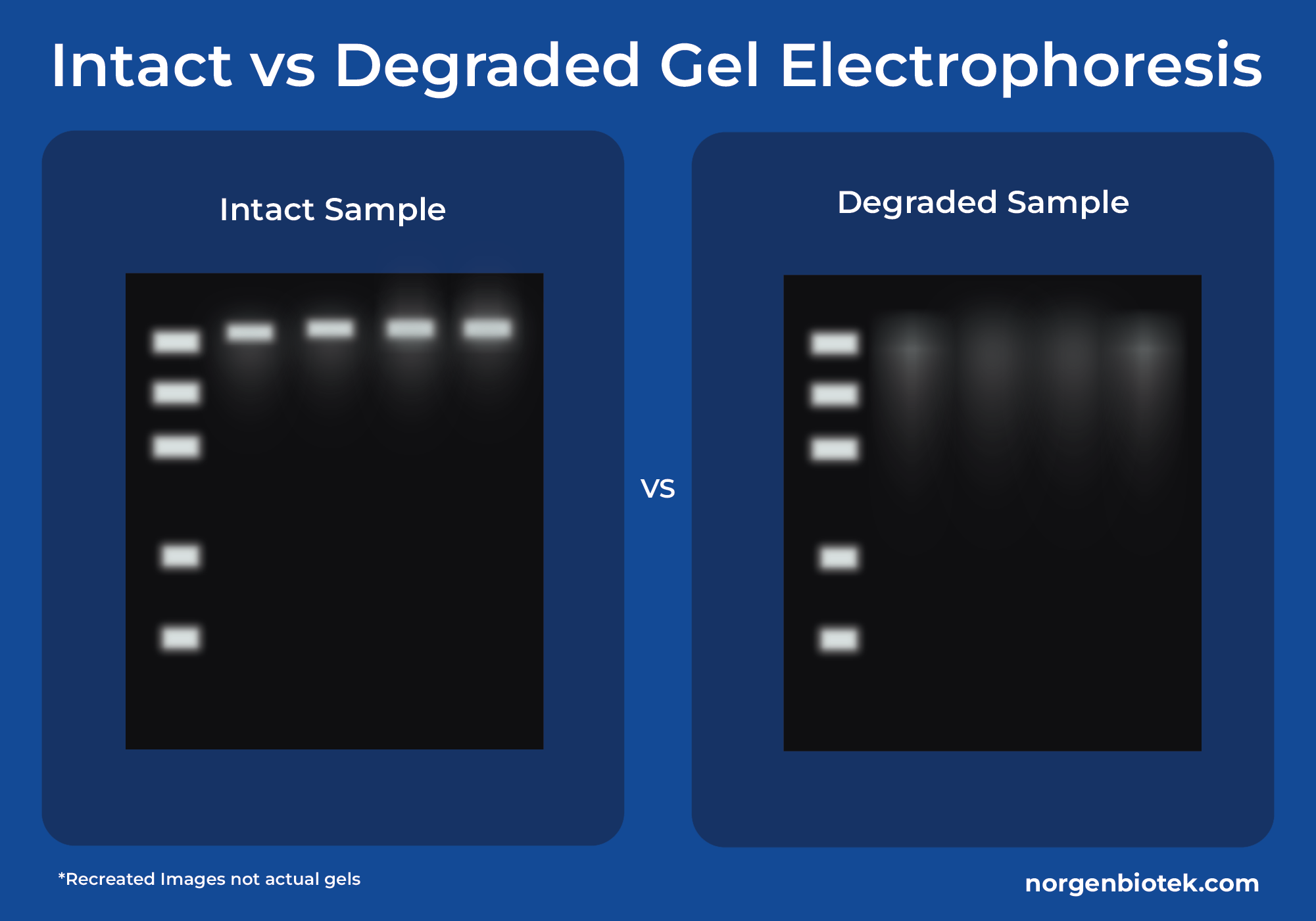
The Battle Between Stool Nucleic Acids and Hydrolytic Enzymes
Nucleic acids in stool samples begin degrading as soon as they are collected. What causes stool DNA and RNA to degrade? Deoxyribonucleases (DNases) are enzymes responsible for cleaving the DNA by hydrolyzing its phosphodiester backbone.5 Similarly, Ribonucleases (RNases) are enzymes that hydrolyze the 3' and 5' phosphodiester linkages in RNA.6 These enzymes are present in the body to regulate immune responses and protect the body against infections among other things. They also break down extracellular DNA and RNA which reduces inflammation and prevents viral infections from spreading.
When fecal matter is excreted, the DNases and RNases continue attacking the DNA and RNA in the sample. A study in 2019 conducted by Zhang et. al. found this causes fast DNA degradation, poor quality of fecal DNA and RNA isolation, and a low success rate of PCR amplification in downstream processes.7 Norgen Biotek's stool nucleic acid products are optimal for stool DNA and RNA preservation, as the chemical preservative inactivates RNases and DNases and provides the user with a safe, non-infectious stool sample.
The Role of Preservation: Preserving a Snapshot
The microbes in the stool continue to grow after the stool is excreted. The problem is that they grow at different rates outside of the body than when inside of the body. This makes it difficult to know the state of the gut microbiome at the time of the stool exiting the body. It is essential to preserve a snapshot of microbial diversity of the stool in order to properly assess the health of the gut. On top of that, stool DNA and RNA are highly sensitive due to the activity of DNases and RNases.

So what are the best conditions to store stool samples to prevent nucleic acid degradation and unwanted microbial growth? Flash freezing at -80°C is the current gold standard due to its ability to prevent DNase and RNase activity and microbial growth, but it is not always a viable option. 8 You may not have a freezer that reaches such low temperatures, you may be in the field where a freezer isn't readily available, and lastly, the stool sample may be subject to fluctuating temperatures during shipment. Well, what is the solution?
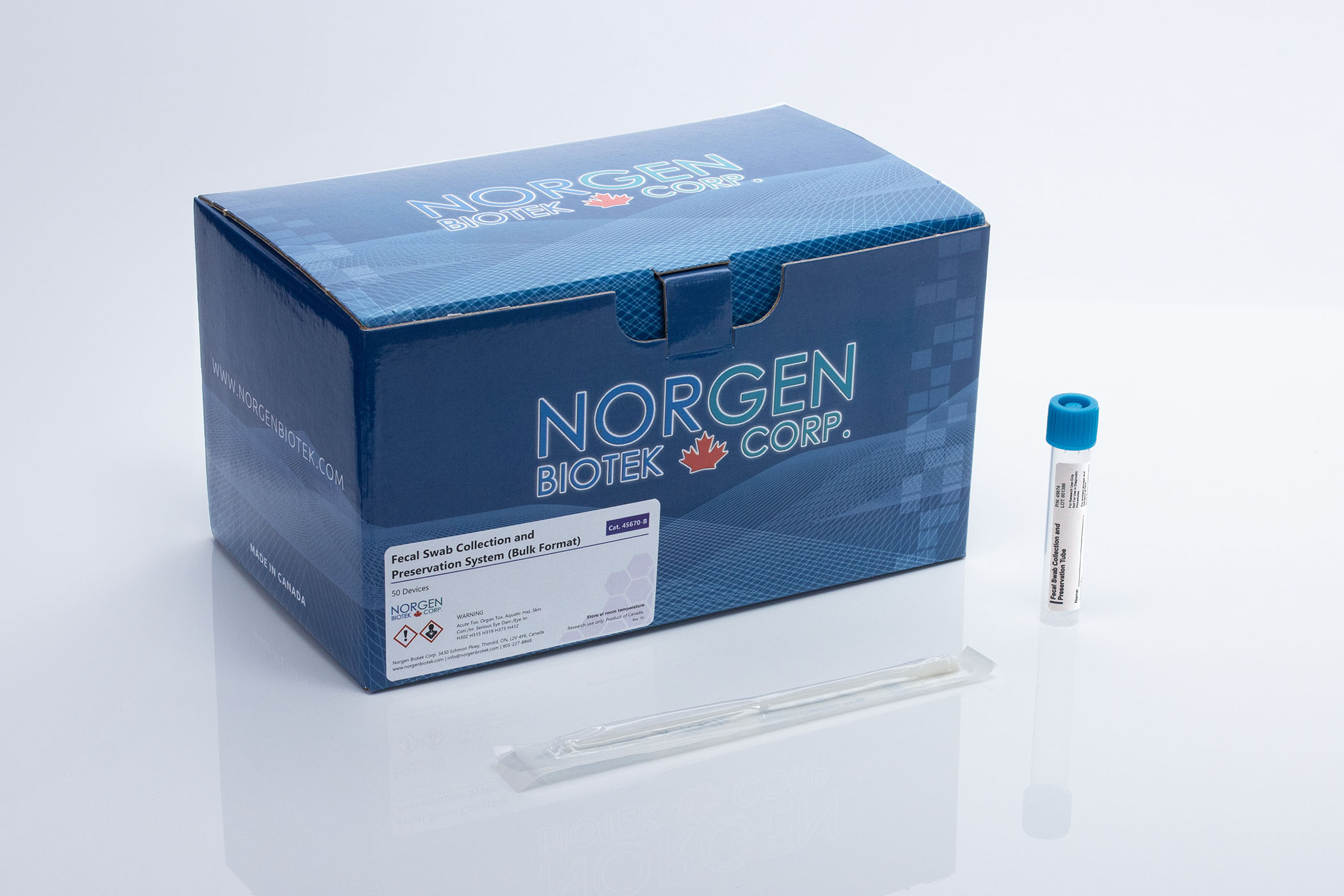
A way to combat that is by eliminating the temperature-sensitive aspect and using Norgen Biotek's stool preservation solutions. Norgen Biotek uses a lytic, chemical preservative that eliminates the growth of microbes in order to preserve the initial composition of the stool while simultaneously stabilizing any free nucleic acid to prevent degradation. At room temperature, the preservative protects stool DNA for up to 2 years and stool RNA for up to 7 days. This makes for easy shipping to processing facilities and stress-free storage. The stool preservative comes in several formats. The most commonly used preservative has the preservative in a tube that is equipped with a scoop for convenient collection. The Stool Nucleic Acid Collection and Preservation System uses the same tube but makes at-home stool collection easy with nitrile gloves and Fe-Col® Collection Paper. There is also the Fecal Swab Collection and Preservation System which, as the name suggests, is designed in a swab format and is commonly used for thin stool samples, as well as for samples collected from infant humans and various mammals. The Fecal DNA Collection & Preservation Mini Tubes are perfect for small pellet samples from small animals and contain pre-loaded beads that eliminate the process of transferring the sample to a different tube for bead beating.
How Pure is your Purified DNA?
PCR testing is a commonly used technique to analyze DNA. Polymerase Chain Reaction (PCR) amplifies the purified stool DNA for better detection of pathogens. PCR is also usually the first step during library preparation for metagenomic sequencing which will be discussed later in this blog.
Purified DNA is often not as pure as you think it is. A 2009 study by Oikarinen et. al. found that PCR inhibitors including bile salts, polysaccharides, proteins, and humic acid are abundant in human stool. These can make their way to the isolated DNA if the purification method is not sufficient in removing these inhibitors, which then interferes with the PCR amplification process. Studies found it can lead to false negative results.9 Including an internal PCR control helps you reduce false negatives, which is why Norgen's Taqman PCR-based pathogen detection kits always include an internal control to ensure accurate results. Furthermore, Norgen's Stool DNA Isolation Kit is optimized to isolate DNA from both the host and microbes which is essential for pathogen detection, while also eliminating PCR inhibitors and other contaminants. This results in high-quality DNA for sensitive downstream applications such as pathogen detection and genomic sequencing.
How Stool DNA Reveals Gut Microbiome Composition
What can we use stool DNA for, and why should we study it? Apart from pathogen detection, fecal DNA can be analyzed to determine the gut microbiome diversity which can help us understand disease correlation. The 16S rRNA gene is a universal genetic marker found in all gut microbes, serving as a critical component of the ribosomal machinery. Its sequence contains highly conserved regions shared among all bacterial species, interspersed with variable regions unique to each species. These sequence variations enable researchers to differentiate and identify microbial species, making the 16S rRNA gene a cornerstone for microbiome profiling and taxonomy. Subjecting bacterial community DNA to NGS (Next Generation Sequencing) targeting the 16S rRNA gene allows the researchers to identify and classify the diverse bacterial populations.
Health conditions, diet, geographical location, and treatments for diseases can alter the microbiome diversity. Research published in 2022 by Righi et. al. studied the impact of COVID-19 and antibiotic treatments on gut microbiome by sequencing 16S rRNA using stool samples from COVID 19-patients between April 2020 and December 2020. The stool samples were collected and preserved at -80°C until nucleic acids were extracted. Then DNA was extracted using Norgen Biotek's Stool DNA Isolation Kit and quantified. Lastly, 16S rRNA sequencing was performed after amplifying the 16S rRNA gene V1-V3 region. This study found that the severity of COVID-19 was positively correlated with an increase in opportunistic bacteria while reducing beneficial bacteria such as Agathobacter in the gut. The severity of COVID-19 was determined by measuring the fraction of inspired oxygen, and inflammatory markers (and other techniques they used in a paper). Agathobacter showed a negative correlation with the fraction of inspired oxygen. on the other hand, they found an abundance of Pseudomonas which is a prevalent opportunistic bacteria associated with the increase in inflammatory markers.10
Another study from 2021 used stool DNA to explore how the gut microbiome affects immunotherapy results in patients with bladder cancer. Cancer cells evade the immune system by overexpressing certain proteins. Immune Checkpoint Inhibitors (ICIs) are a type of cancer treatment that blocks these proteins, allowing the immune system to kill cancer cells. The researchers used Norgen Biotek's Stool Nucleic Acid Collection and Preservation Tubes to collect stool samples from 42 patients. They extracted DNA from the stool samples using Norgen Biotek's Stool DNA Isolation Kit to perform 16S sequencing. They found that the 23 patients who responded to the ICIs treatment and it was discovered they had a higher diversity of gut microbiota compared to those who did not respond to the treatment. They concluded the gut microbiota was linked to how well the patient responded to the ICI treatment. 11 According to the studies discussed above and many other studies, stool DNA can help us investigate microbial diversity or genetic variation in host cells. However, to understand functional pathways active in the microbes as well as host cells, we need to study stool RNA.
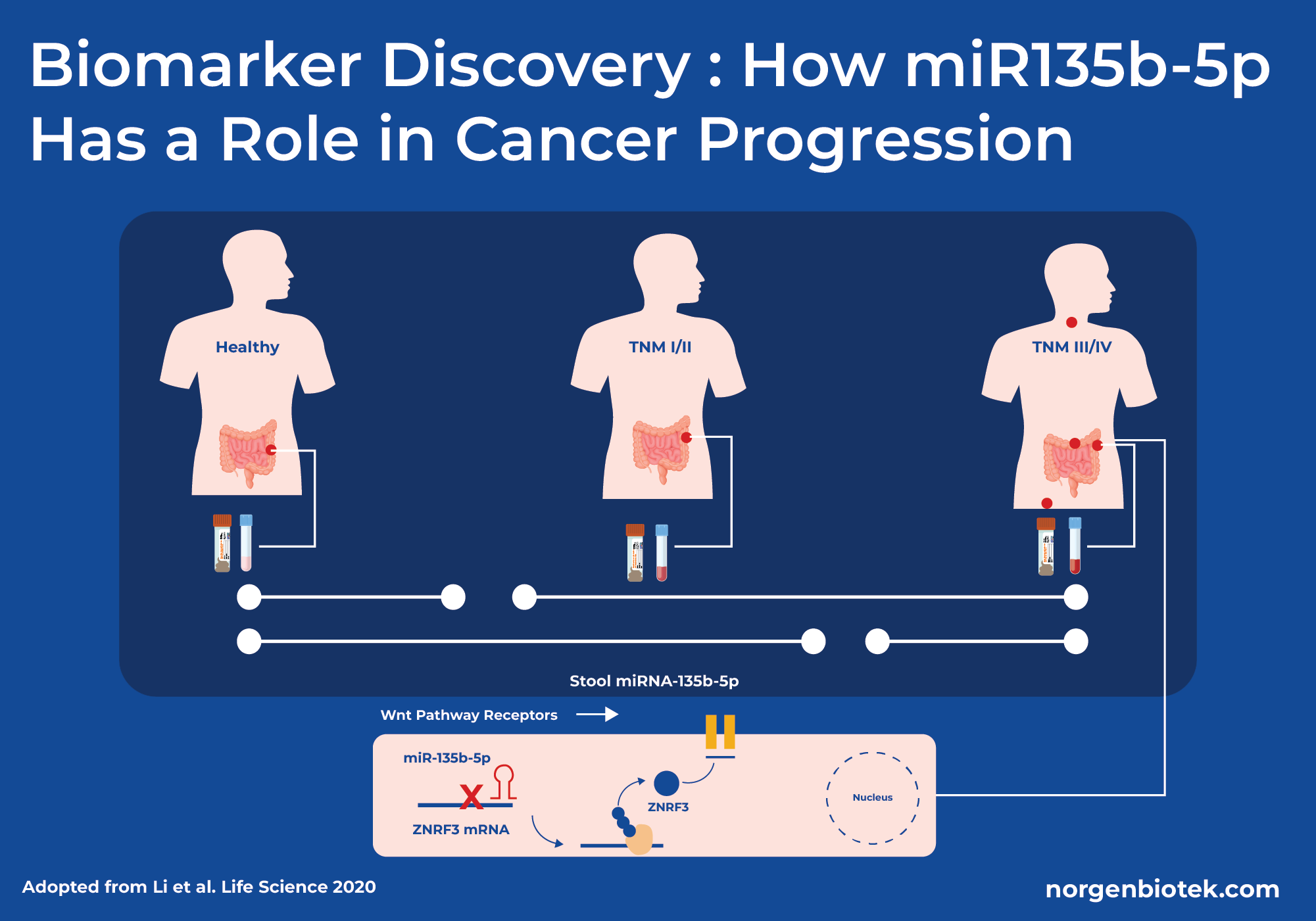
Stool RNA as a Key to Cancer Biomarker Discovery
Stool RNA analysis provides a broad view of gene expression, including insights into microbial activity and host responses. Diving deeper, stool small RNA analysis focuses specifically on smaller RNA molecules, such as microRNAs (miRNAs), which play critical regulatory roles in gene expression and offer unique information about host-microbiome interactions and disease processes. These small molecules show significant potential as biomarkers for many diseases including cancers, and stool miRNA can be a great tool for detecting colorectal cancer. miRNAs are one of the smallest types of RNA and they require highly optimized isolation methods. Fortunately, Norgen's proprietary Silicon Carbide technology isolates high-quality miRNA as well as large RNA with similar efficiency and purity, ensuring reliable results for downstream applications.
A study in 2020 explored the identification of host miRNA (miR-135b-5p) from stool samples as a non-invasive biomarker for colorectal cancer. The study included 77 colorectal cancer patients and 29 healthy control patients and they used Norgen Biotek's Stool Total RNA Purification kit to isolate fecal RNA. The research team then conducted a quantitative real-time PCR for miR-135b-5p. The study found miR-135b-5p to be upregulated in patients with colorectal cancer and it can distinguish between healthy individuals and colorectal cancer patients. The upregulation of miR-135b-5p effectively reduces the protein expression levels of the ZNRF3 gene. The ZNRF3 gene is responsible for inhibiting WNT pathways which is crucial for cell growth and differentiation. The downregulation of ZNRF3 protein expression increases WNT signaling which causes faster spread of cancer. Overall, the research team concluded miR-135b-5p isolated from stool samples to be a promising biomarker for detecting colorectal cancer in patients with TNM (Tumor, Node, Metastasis) stage III or IV.12
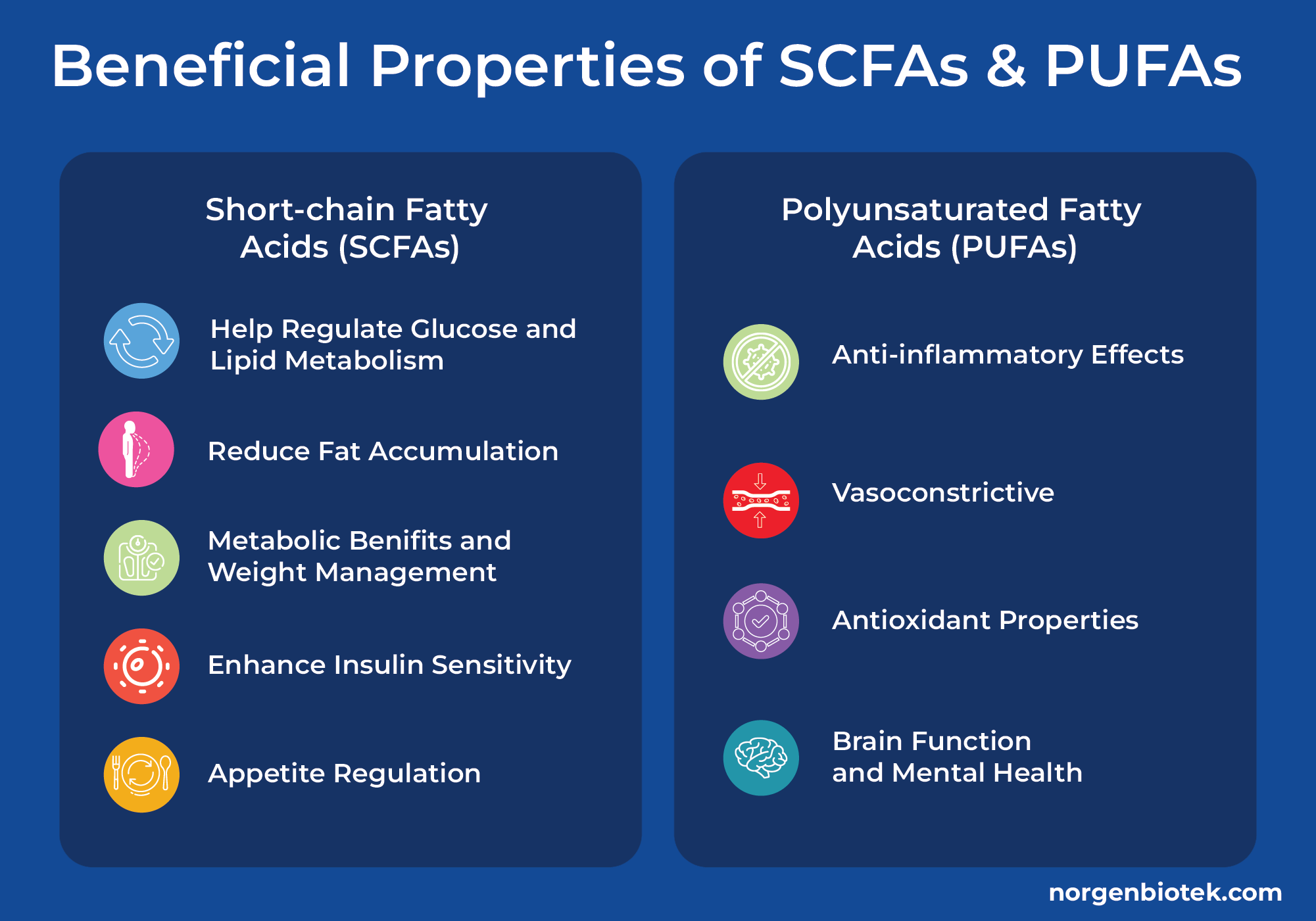
Metabolomic Marvel: What More Can Stool Tell Us?
Did you know stool samples can be used to analyze short-chain fatty acids (SCFAs), polyunsaturated fatty acids (PUFAs), and many other metabolites? SCFAs are produced in the large intestine by bacteria that convert indigestible polysaccharides into acetate, propionate, butyrate, and more. SCFAs play a crucial role in mucus production and maintaining the integrity of the intestinal barrier.14 SCFAs, particularly propionate, have been linked to improved metabolic health. They can help regulate glucose and lipid metabolism,16 reduce fat accumulation,17 and enhance insulin sensitivity.18 Some studies suggest that SCFAs may also play a role in appetite regulation, potentially helping with weight management.19 PUFAs on the other hand are not produced in the gut but are obtained from foods with healthy fats such as oily fish and nuts. Omega-3 and Omega-6 are the most common types of PUFAs that are known for their health benefits such as their antiinflammatory, vasoconstrictive, and antioxidant properties.15
In 2023, Liam O'Neill et. al. explored the effects of milkfat consumption on the gut microbiome of bariatric surgery patients. They studied the gut microbiome and the metabolites in the gut. The research team used Norgen Biotek's Stool Nucleic Acid Collection and Preservation Tubes to collect stool from 9 patients 2 weeks before surgery, at the time of surgery, and 1, 3, and 6 months after surgery. Because Norgen's preservative is also compatible with metabolomic studies, they were able to perform gas chromatography-mass spectrometry (GC-MS) for non-targeted fatty acid analysis of 30 fatty acids. The study found an increase in bacteria such as Roseburia, Christensenellaceae, Blautia, and other Bacteroides in patients who consume high amounts of milk fat. These bacteria are associated with weight loss, for example, according to the study, “Blautia is proposed to decrease obesity by producing butyric acid and acetic acid”. They also discovered that “three patients showing higher concentrations of saturated fatty acids such as myristic acid were low-milk-fat consumers. On the other hand, a high-milk-fat consumer showed a very high concentration of beneficial omega-3-polyunsaturated fatty acids”. They concluded that the consumption of milk fat increases the presence of polyunsaturated fatty acids and weight-loss-associated bacteria in the gut.13
Overall, stool samples play a vital role in a wide range of detection, diagnostic, and analytical applications. From pathogen identification to cancer detection, stool offers valuable insights into the host's overall health. Ensuring that stool samples are properly collected and preserved is critical for capturing an accurate snapshot of the gut microbiome. At Norgen Biotek, we are committed to empowering researchers and clinicians with high-quality solutions for every step of the stool analysis process. From collection and preservation to nucleic acid extraction and TaqMan PCR detection, our products are designed to deliver reliable, accurate results.
Discover how Norgen's TaqMan-based PCR kits, 16S and ITS library prep kits, and NGS services can elevate your research and diagnostic workflows. Partner with us to unlock the full potential of your stool analysis journey. Together, let's advance the science of health and discovery.