The Science Behind Highly Sensitive Cell-Free DNA Isolation
Quality Samples = Quality Results
Isaac Newton famously said, “If I have seen further it is by standing on the shoulders of giants.” However, what if said giants weren't looking in the right direction? According to a recent publication, "32%-75% of all testing errors are the result of issues that arise during the pre-analytical phase".1 Unquestionably, this is due to a lack of regulated protocols for sample collection and handling. After all, only quality samples can generate quality results.
As the push towards the utilization of cell-free DNA biomarkers in precision medicine increases, more research is bringing attention to the need for standardized sample handling protocols within the field. Cell-free DNA is highly fragmented and the majority (90%) are found in the range between 80 bp-200 bp.2 To make things more complicated, urine samples are highly variable among individuals and are much more diluted compared to other samples like blood. There is no need to panic though, as we’ve got some helpful tips for working with urine!
Although the clinical application of cell-free DNA (and cell-free RNA) within urine shows great promise in oncology, it remains a challenging sample type to work with due to a lack of standardized pre-analytical protocols. Urine has evidently been shown to be a source of biomarkers, however, due to its nature as a highly diluted cell-free sample, it requires careful consideration during the collection procedure. Compared to traditional tissue biopsies, urine liquid biopsy offers many advantages, including the non-invasive detection of circulating fetal DNA for prenatal genetic diagnostics and use as a potential tool to capture tumor heterogeneity in metastatic cancer patients.2
In addition to sample collection, sensitive methods for nucleic acid isolation and quantification are areas which require standardization. All steps that have the potential to affect the outcome and informational value of the performed analyses should follow a standardized protocol since they are used for the discovery of novel biomarkers.
NORBLOG
Want to hear more from Norgen?
Join over 10,000 scientists, bioinformaticians, and researchers who receive our exclusive deals, industry updates, and more, directly to their inbox.
For a limited time, subscribe and SAVE 10% on your next purchase!
SIGN UP
A Need for Standardization
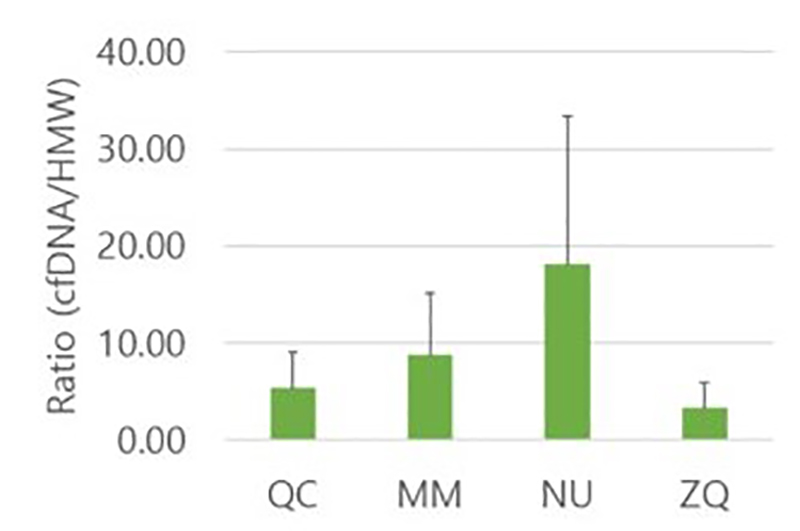
In a 2020 publication by Lee et al., a group of scientists compared the efficiencies of four commercial kits for urinary cell-free DNA (cf-DNA) isolation. This research was an effort to bridge the gap on the lack of standardized isolation methods for small DNA fragments that are critical when investigating urologic malignancies.3 The group demonstrated that Norgen’s Urine Cell-Free Circulating DNA Purification Kit was the most efficient for isolation of more fragmented cf-DNA in the range of 50–100 bp with the lowest cellular genomic DNA contamination (Figure 1).3
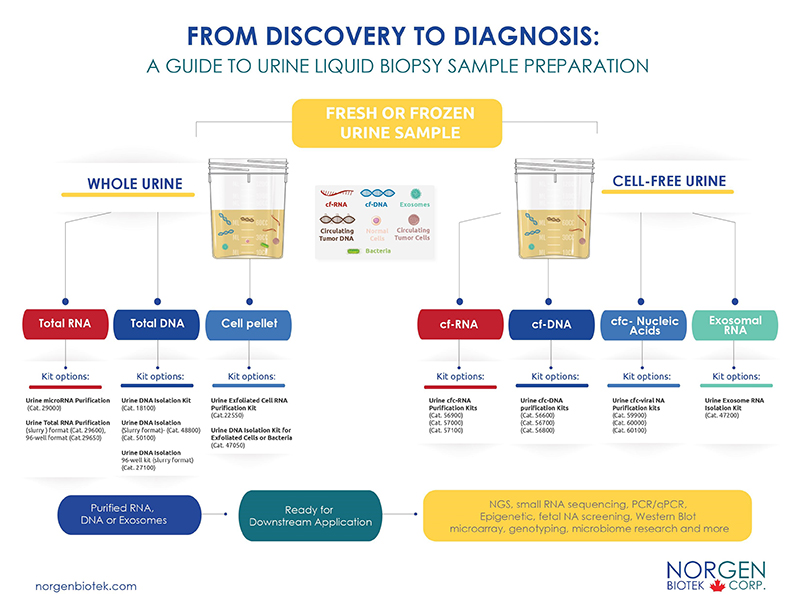
Norgen has since developed a convenient workflow to assist researchers in selecting the most suitable kits for their projects. Whether you are working with frozen or fresh samples, we offer a convenient method to help you isolate the analyte of choice (RNA, DNA, exosome) with the highest efficiency using our silicon carbide technology. Silicon carbide ensures that the smallest RNA and DNA fragments are isolated and compatible for a variety of sensitive downstream applications. Let us help you eliminate pre-analytical testing errors and assist you in reaching your research goals!
-
Szilágyi M, Pös O, Márton É, et al. Circulating cell-free nucleic acids: Main characteristics and clinical application. International Journal of Molecular Sciences.2020;21(18):6827. doi:10.3390/ijms21186827
-
Augustus E, Van Casteren K, Sorber L, et al. The art of obtaining a high yield of cell-free DNA from urine. PLOS ONE.2020;15(4). doi:10.1371/journal.pone.0231058
-
Lee EY, Lee E-J, Yoon H, Lee DH, Kim KH. Comparison of four commercial kits for isolation of urinary cell-free DNA and sample storage conditions. Diagnostics.2020;10(4):234doi:10.3390/diagnostics10040234