Maintaining Microbiome Composition with Superior Preservation
Containing trillions of microorganisms such as bacteria, viruses, and fungi, the gut microbiome contains the largest population of microbes in the human body.1 Governing such a vast community of microorganisms comes with great responsibility, as the interactions between the host and the microbiota can significantly influence ones metabolism, neurological function2, inflammation, and cellular and immune responses.3 However, great responsibility comes with great power- the disruption of this relationship can give rise to a plethora of problems such as inflammatory bowel disease4, obesity5, cancer6, cardiovascular disease, allergies, and diabetes.7
The Importance of Sample Preservation
Over the past two decades, scientists have recognized the importance of studying the gut microbiome and its substantial effect on human physiology. This has been made possible through the collection of stool samples, which is a non-invasive sampling method that enables us to accurately profile the microbiota composition. However, no two profiles are the same, as the abundance and variety in microorganisms found in the gastrointestinal system are largely diverse and heavily influenced by a person's environment, lifestyle, diet, and genetics. Consequently, microbiome studies have evolved to encompass a greater demographic. Since the majority of people don't live near a laboratory, stool samples typically require shipping for an extended period of time. However, stool samples, in particular, often experience nucleic acid degradation and bacterial growth which alters the sample by the time it arrives for testing. For many years, freezing was considered the gold standard for sample preservation, but it was proven to be costly and difficult to upkeep with at-home collection, especially when dealing with warmer climate countries. Chemical preservation of samples was eventually discovered as an alternative to freezing and cold-chain transport, allowing for storage at ambient temperature as well as viral, fungal, and bacterial inactivation.
Freezing vs. Chemical Sample Preservation
 Since collection and storage conditions can introduce significant bias and
variation, determining the best preservation method for microbiome stability
from point-of-collection is crucial - and that is exactly what a research
group in Hong Kong set out to do. Chen et al. (2019) explored the preservation
capabilities of six popular chemical preservation solutions (Norgen, OMNI,
RNAlater, CURNA, HEMA, and Shield) and compared them to immediate freezing at
-80°C.8 Stool samples were taken from 5 individuals and were
aliquoted for storage in the aforementioned preservation solutions for 7 days
at room temperature with the intention of mimicking shipping conditions. The
samples underwent total DNA extraction utilizing a bead-beating method,
followed by amplicon
sequencing
targeting three hypervariable regions of the
16S rRNA gene
(V1-V2, V3-V4, and V-4) in aims to additionally explore the implications of
primer sensitivity and their ability to distinguish specific bacterial
taxa.8
Since collection and storage conditions can introduce significant bias and
variation, determining the best preservation method for microbiome stability
from point-of-collection is crucial - and that is exactly what a research
group in Hong Kong set out to do. Chen et al. (2019) explored the preservation
capabilities of six popular chemical preservation solutions (Norgen, OMNI,
RNAlater, CURNA, HEMA, and Shield) and compared them to immediate freezing at
-80°C.8 Stool samples were taken from 5 individuals and were
aliquoted for storage in the aforementioned preservation solutions for 7 days
at room temperature with the intention of mimicking shipping conditions. The
samples underwent total DNA extraction utilizing a bead-beating method,
followed by amplicon
sequencing
targeting three hypervariable regions of the
16S rRNA gene
(V1-V2, V3-V4, and V-4) in aims to additionally explore the implications of
primer sensitivity and their ability to distinguish specific bacterial
taxa.8
The researchers determined first and foremost that the most significant contributor to substantial changes in gut microbial compositions was due to inter-subject variation. This underlines the need for widespread microbiome studies while highlighting the significance of proper preservation in order to reduce variations in sample composition between subjects. The authors noted that preservation methods have the ability to introduce changes in the microbiota profile which inflict bias for downstream applications.
Superior Sample Preservation
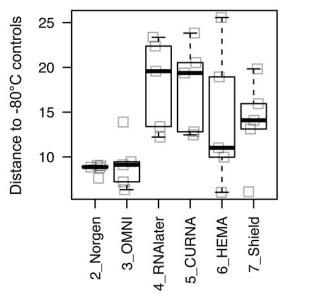
Through the use of bioinformatics, weighted UniFrac distances between two pairwise points were generated between the chemically preserved and standard frozen samples, as seen in figure 1. From the 6 preservation solutions that were compared, the Norgen and OMNI preservatives stood out from the rest with the smallest mean distance to the standard frozen samples, signifying the least shift in community composition.
NORBLOG
Want to hear more from Norgen?
Join over 10,000 scientists, bioinformaticians, and researchers who receive our exclusive deals, industry updates, and more, directly to their inbox.
For a limited time, subscribe and SAVE 10% on your next purchase!
SIGN UP
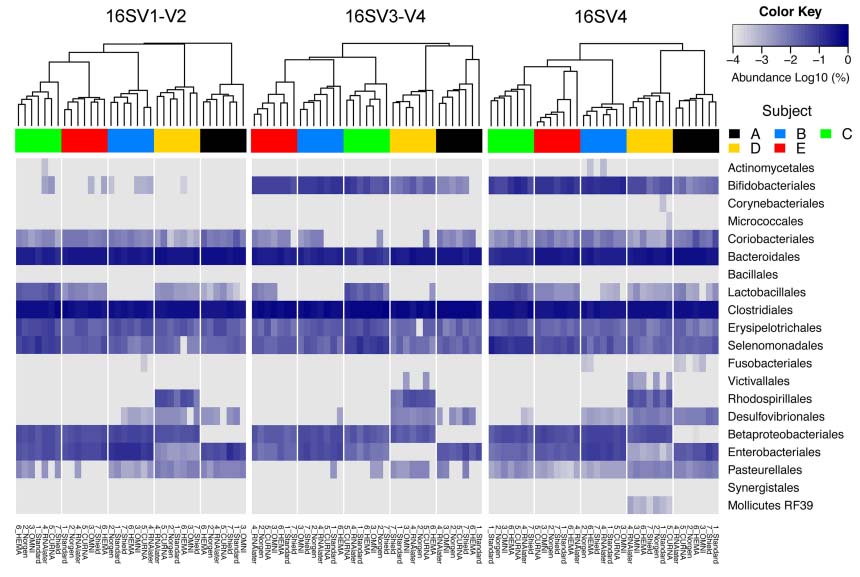
Furthermore, the authors determined that different microbial community profiles are generated based on the primer pairs chosen to target different parts of the bacterial 16S rRNA gene, as seen in figure 2. However, "...samples in Norgen preservative consistently showed the least shift in community composition relative to -80°C standards"8, regardless of the region targeted. Alternatively, samples stored in Shield reported a significant increase in the relative abundance of Faecalibacterium compared to the standard, while OMNI presented a decrease in relative abundance compared to the -80°C of Lachnospiraceae.8
The authors also evaluated the bactericidal ability of the preservatives by observing any microbial growth under aerobic and anaerobic conditions. They determined that only Norgen, OMNI, HEMA, and Shield immediately rendered the samples noninfectious and inhibited bacterial growth. This makes the samples safe to transport and handle without risk of infection.
Overall, the rising awareness of the importance of microbiome research creates an urge for a safe, reliable, and effective mode of sample preservation. As indicated in the study above, Norgen's Total Nucleic Acid Collection and Preservation Tubes check all the boxes, from minimizing shifts in microbial profiles to facilitating safe and stable stool transportation.
If you'd like to try Norgen's Total Nucleic Acid Preservation Tubes in your microbial workflow, request a sample and we'll send you a free device to test out!
Related Products and Services for Gut Microbiome Research
-
Lloyd-Price, J., Abu-Ali, G. & Huttenhower, C. The healthy human microbiome. Genome Med 8, 51 (2016). https://doi.org/10.1186/s13073-016-0307-y
-
Slingerland A.E., Stein-Thoeringer C.K. (2018) Microbiome and Diseases: Neurological Disorders. In: Haller D. (eds) The Gut Microbiome in Health and Disease. Springer, Cham.https://doi.org/10.1007/978-3-319-90545-7_18
-
Zitvogel, L., Daillère, R., Roberti, M. et al. Anticancer effects of the microbiome and its products. Nat Rev Microbiol 15, 465-478 (2017). https://doi.org/10.1038/nrmicro.2017.44
-
Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843-54.
-
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480-4.
-
Schwabe, R., Jobin, C. The microbiome and cancer. Nat Rev Cancer 13, 800-812 (2013). https://doi.org/10.1038/nrc3610
-
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55-60.
-
Chen, Z., Hui, P.C., Hui, M., Yeoh Y.K., Wong, P.Y,. Chan, M.C.W., Wong, M.C.S., Ng, S.C., Chan, F.K.L., Chan, P.K.S. (2019). Impact of Preservation Method and 16S rRNA Hypervariable Region on Gut Microbiota Profiling.